Abstract
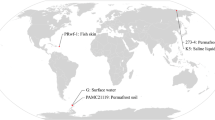
Bacteria of the genus Exiguobacterium are low G + C, Gram-positive facultative anaerobes that have been repeatedly isolated from ancient Siberian permafrost. In addition, Exiguobacterium spp. have been isolated from markedly diverse sources, including Greenland glacial ice, hot springs at Yellowstone National Park, the rhizosphere of plants, and the environment of food processing plants. Strains of this hereto little known bacterium that have been retrieved from such different (and often extreme) environments are worthy of attention as they are likely to be specifically adapted to such environments and to carry variations in the genome which may correspond to psychrophilic and thermophilic adaptations. However, comparative genomic investigations of Exiguobacterium spp. from different sources have been limited. In this study, we employed different molecular approaches for the comparative analysis of 24 isolates from markedly diverse environments including ancient Siberian permafrost and hot springs at Yellowstone National Park. Pulsed-field gel electrophoresis (PFGE) with I-CeuI (an intron-encoded endonuclease), AscI and NotI were optimized for the determination of genomic fingerprints of nuclease-producing isolates. The application of a DNA macroarray for 82 putative stress-response genes yielded strain-specific hybridization profiles. Cluster analyses of 16S rRNA gene sequence data, PFGE I-CeuI restriction patterns and hybridization profiles suggested that Exiguobacterium strains formed two distinct divisions that generally agreed with temperature ranges for growth. With few exceptions (e.g., Greenland ice isolate GIC31), psychrotrophic and thermophilic isolates belonged to different divisions.




Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Anderson CR, Cook GM (2004) Isolation and characterization of arsenate-reducing bacteria from arsenic-contaminated sites in New Zealand. Curr Microbiol 48:341–347
Bogdanova ES, Bass IA, Minakhin LS, Petrova MA, Mindlin SZ, Volodin AA, Kalyaeva ES, Tiedje JM, Hobman JL, Brown NL, Nikiforov VG (1998) Horizontal spread of mer operons among Gram-positive bacteria in natural environments. Microbiology 144:609–620
Bogdanova E, Minakhin L, Bass I, Volodin A, Hobman JL, Nikiforov V (2001) Class II broad-spectrum mercury resistance transposons in Gram-positive bacteria from natural environments. Res Microbiol 152:503–514
Chaturvedi P, Shivaji S (2006) Exiguobacterium indicum sp nov., a psychrophilic bacterium from the Hamta glacier of the Himalayan mountain ranges of India. Int J Syst Evol Microbiol 56:2765–2770
Chaturvedi P, Prabahar V, Manorama R, Pindi PK, Bhadra B, Begum Z, Shivaji S (2008) Exiguobacterium soli sp nov., a psychrophilic bacterium from the McMurdo Dry Valleys, Antarctica. Int J Syst Evol Microbiol 58:2447–2453
Collins MD, Kroppenstedt RM (1983) Lipid-composition as a guide to the classification of some coryneform bacteria-containing an A4-alpha type peptidoglycan. Syst Appl Microbiol 4:95–104
Collins MD, Lund BM, Farrow JAE, Schleifer KH (1983) Chemotaxonomic study of an alkaliphilic bacterium, Exiguobacterium aurantiacum gen nov., sp. nov. J Gen Microbiol 129:2037–2042
Corkill JE, Graham R, Hart CA, Stubbs S (2000) Pulsed-field gel electrophoresis of degradation-sensitive DNAs from Clostridium difficile PCR ribotype 1 strains. J Clin Microbiol 38:2791–2792
Crapart S, Fardeau ML, Cayol JL, Thomas P, Sery C, Ollivier B, Combet-Blanc Y (2007) Exiguobacterium profundum sp nov., a moderately thermophilic, lactic acid-producing bacterium isolated from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol 57:287–292
Farrow JAE, Wallbanks S, Collins MD (1994) Phylogenetic interrelationships of round-spore-forming bacilli containing cell-walls based on lysine and the non-spore-forming genera Caryophanon, Exiguobacterium, Kurthia, and Planococcus. Int J Syst Bacteriol 44:74–82
Fruhling A, Schumann P, Hippe H, Straubler B, Stackebrandt E (2002) Exiguobacterium undae sp nov. and Exiguobacterium antarcticum sp. nov. Int J Syst Evol Microbiol 52:1171–1176
Gibson JR, Sutherland K, Owen RJ (1994) Inhibition of DNase activity in PFGE analysis of DNA from Campylobacter jejuni. Lett Appl Microbiol 19:357–358
Hara I, Ichise N, Kojima K, Kondo H, Ohgiya S, Matsuyama H, Yumoto I (2007) Relationship between the size of the bottleneck 15 angstrom from iron in the main channel and the reactivity of catalase corresponding to the molecular size of substrates. Biochemistry 46:11–22
Higgins DG, Sharp PM (1988) Clustal—a package for performing multiple sequence alignment on a microcomputer. Gene 73:237–244
Hwang BY, Kim JH, Kim J, Kim BG (2005) Screening of Exigubacterium acetylicum from soil samples showing enantioselective and alkalitolerant esterase activity. Biotechnol Bioprocess Eng 10:367–371
Ito K (2005) Ribosome-based protein folding systems are structurally divergent but functionally universal across biological kingdoms. Mol Microbiol 57:313–317
Jeffries CD, Holtman DF, Guse DG (1957) Rapid method for determining the activity of microorganisms on nucleic acids. J Bacteriol 73:590–591
Kasana RC, Yadav SK (2007) Isolation of a psychrotrophic Exiguobacterium sp. SKPB5 (MTCC 7803) and characterization of its alkaline protease. Curr Microbiol 54:224–229
Kim IG, Lee MH, Jung SY, Song JJ, Oh TK, Yoon JH (2005) Exiguobacterium aestuarii sp nov. and Exiguobacterium marinum sp. nov., isolated from a tidal flat of the Yellow Sea in Korea. Int J Syst Evol Microbiol 55:885–889
King GM, Stetzenbach LD, Klima-Comba A (2005) Analysis of cultivable airborne bacteria from an altitude gradient on Kilauea and Mauna Loa Volcanoes (Hawaii). In: ASM general 105th meeting, ASM Press, Atlanta, GA, pp N-094
Klaassen CHW, van Haren HA, Horrevorts AM (2002) Molecular fingerprinting of Clostridium difficile isolates: Pulsed-field gel electrophoresis versus amplified fragment length polymorphism. J Clin Microbiol 40:101–104
Knudston KE, Haas EJ, Iwen PC, Ramaley WC, Ramaley RF (2001) Characterization of a Gram-positive, non-spore-forming Exiguobacterium-like organism isolated from a Western Colorado (USA) hot spring. In: ASM general 101th meeting, ASM Press, Orlando, FL, pp I-92
Kumar A, Singh V, Kumar R (2006) Characterization of an alkaliphile, Exiguobacterium sp. and it’s application in bioremediation. In: International conference on extremophiles, Brest, France, p L89
Liu SL, Schryvers AB, Sanderson KE, Johnston RN (1999) Bacterial phylogenetic clusters revealed by genome structure. J Bacteriol 181:6747–6755
Lopez L, Pozo C, Rodelas B, Calvo C, Juarez B, Martinez-Toledo MV, Gonzalez-Lopez J (2005) Identification of bacteria isolated from an oligotrophic lake with pesticide removal capacities. Ecotoxicology 14:299–312
Lopez-Cortes A, Schumann P, Pukall R, Stackebrandt E (2006) Exiguobacterium mexicanum sp nov. and Exiguobacterium artemiae sp. nov., isolated from the brine shrimp Artemia franciscana. Syst Appl Microbiol 29:183–190
Maidak BL, Cole JR, Lilburn TG, Parker CT, Saxman PR, Farris RJ, Garrity GM, Olsen GJ, Schmidt TM, Tiedje JM (2001) The RDP-II (Ribosomal Database Project). Nucleic Acids Res 29:173–174
Miteva VI, Sheridan PP, Brenchley JE (2004) Phylogenetic and physiological diversity of microorganisms isolated from a deep Greenland glacier ice core. Appl Environ Microbiol 70:202–213
Newnham E, Chang N, Taylor DE (1996) Expanded genomic map of Campylobacter jejuni UA580 and localization of 23S ribosomal rRNA genes by I-CeuI restriction endonuclease digestion. FEMS Microbiol Lett 142:223–229
Okeke BC, Laymon J, Oji C, Crenshaw S (2007) Rapid bioreduction of hexavalent chromium in water by Exiguobacterium sp. GS1. In: ASM general 107th meeting, ASM Press, Toronto, ON, Canada, pp Q-199
Pattanapipitpaisal P, Mabbett AN, Finlay JA, Beswick AJ, Paterson-Beedle M, Essa A, Wright J, Tolley MR, Badar U, Ahmed N, Hobman JL, Brown NL, Macaskie LE (2002) Reduction of Cr(VI) and bioaccumulation of chromium by Gram-positive and Gram-negative microorganisms not previously exposed to Cr-stress. Environ Technol 23:731–745
Petrova MA, Mindlin SZ, Gorlenko ZM, Kalyaeva ES, Soina VS, Bogdanova ES (2002) Mercury-resistant bacteria from permafrost sediments and prospects for their use in comparative studies of mercury resistance determinants. Russ J Genet 38:1330–1334
Ponder MA, Gilmour SJ, Bergholz PW, Mindock CA, Hollingsworth R, Thomashow MF, Tiedje JM (2005) Characterization of potential stress responses in ancient Siberian permafrost psychroactive bacteria. FEMS Mcirobiol Ecol 53:103–115
Qiu YH, Kathariou S, Lubman DM (2006) Proteomic analysis of cold adaptation in a Siberian permafrost bacterium—Exiguobacterium sibiricum 255–15 by two-dimensional liquid separation coupled with mass spectrometry. Proteomics 6:5221–5233
Regha K, Satapathy AK, Ray MK (2005) RecD plays an essential function during growth at low temperature in the Antarctic bacterium Pseudomonas syringae Lz4W. Genetics 170:1473–1484
Reiter B, Pfeifer U, Schwab H, Sessitsch A (2002) Response of endophytic bacterial communities in potato plants to infection with Erwinia carotovora subsp. atroseptica. Appl Environ Microbiol 68:2261–2268
Rodrigues DF, Tiedje JM (2007) Multi-locus real-time PCR for quantitation of bacteria in the environment reveals Exiguobacterium to be prevalent in permafrost. FEMS Mcirobiol Ecol 59:489–499
Rodrigues DF, Goris J, Vishnivetskaya T, Gilichinsky D, Thomashow MF, Tiedje JM (2006) Characterization of Exiguobacterium isolates from the Siberian permafrost Description of Exiguobacterium sibiricum sp. nov. Extremophiles 10:285–294
Rodrigues DF, Ivanova N, He Z, Huebner M, Zhou J, Tiedje JM (2008) Architecture of thermal adaptation in an Exiguobacterium sibiricum strain isolated from 3 million year old permafrost: a genome and transcriptome approach. BMC Genomics 9:547
Rozen S, Skaletsky H (2000) Primer 3 on the WWW for general users and for biologist programmers. Bioinformatics methods and protocols: methods in molecular biology. In: Krawetz S, Misener S (eds). Humana Press, Totowa, NJ, pp 365–386
Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M, Barrell B (2000) Artemis: sequence visualization and annotation. Bioinformatics 16:944–945
Saitou N, Nei M (1987) The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Suga S, Koyama N (2000) Purification and properties of a novel azide-sensitive ATPase of Exiguobacterium aurantiacum. Arch Microbiol 173:200–205
Suzuki Y, Haruki M, Takano K, Morikawa M, Kanaya S (2004) Possible involvement of an FKBP family member protein from a psychrotrophic bacterium Shewanella sp. SIB1 in cold-adaptation. Eur J Biochem 271:1372–1381
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA 101:11030–11035
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Uma S, Jadhav RS, Kumar GS, Shivaji S, Ray HK (1999) A RNA polymerase with transcriptional activity at 0 degrees C from the Antarctic bacterium Pseudomonas syringae. FEBS Lett 453:313–317
Usuda Y, Kawasaki H, Shimaoka M, Utagawa T (1998) Molecular characterization of guanosine kinase gene from a facultative alkaliphile, Exiguobacterium aurantiacum ATCC 35652. Biochim Biophys Acta-Gene Struct Expr 1442:373–379
Vishnivetskaya TA, Kathariou S (2005) Putative transposases conserved in Exiguobacterium isolates from ancient Siberian permafrost and from contemporary surface habitats. Appl Environ Microbiol 71:6954–6962
Vishnivetskaya TA, Ramaley R, Rodrigues DF, Tiedje JM, Kathariou S (2005) Exiguobacterium from frozen subsurface sediments (Siberian permafrost) and from other sources have growth temperature ranges reflective of the environmental thermocline of their origin. In: The joint international symposia for subsurface microbiology (ISSM 2005) and environmental biogeochemistry (ISEB XVII), Jackson Hole, WY, p 254
Vishnivetskaya TA, Petrova MA, Urbance J, Ponder M, Moyer CL, Gilichinsky DA, Tiedje JM (2006) Bacterial community in ancient Siberian permafrost as characterized by culture and culture-independent methods. Astrobiology 6:400–414
Vishnivetskaya TA, Siletzky R, Jefferies N, Tiedje JM, Kathariou S (2007) Effect of low temperature and culture media on the growth and freeze-thawing tolerance of Exiguobacterium strains. Cryobiology 54:234–240
Vishnivetskaya T, Podar M, Kathariou S, Tiedje JM (2008) Exiguobacterium: the psychrophilic vs the thermophilic lifestyle. In: The 3th international conference on polar and alpine microbiology, Banff, Canada, pp S4–S5
Wada M, Yoshizumi A, Furukawa Y, Kawabata H, Ueda M, Takagi H, Nakamori S (2004) Cloning and overexpression of the Exiguobacterium sp. F42 gene encoding a new short chain dehydrogenase, which catalyzes the stereoselective reduction of ethyl 3-oxo-3-(2-thienyl)propanoate to ethyl (S)-3-hydroxy-3-(2-thienyl)propanoate. Biosci Biotechnol Biochem 68:1481–1488
Yarza P, Richter M, Peplies J, Euzeby J, Amann R, Schleifer KH, Ludwig W, Glockner FO, Rossello-Mora R (2008) The All-Species Living Tree project: A 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol 31:241–250
Yumoto I, Hishinuma-Narisawa M, Hirota K, Shingyo T, Takebe F, Nodasaka Y, Matsuyama H, Hara I (2004) Exiguobacterium oxidotolerans sp nov., a novel alkaliphile exhibiting high catalase activity. Int J Syst Evol Microbiol 54:2013–2017
Zheng SP, Ponder MA, Shih JY, Tiedje JM, Thomashow MF, Lubman DM (2007) A proteomic analysis of Psychrobacter arcticus 273-4 adaptation to low temperature and salinity using a 2-D liquid mapping approach. Electrophoresis 28:467–488
Acknowledgments
We are grateful to R. Ramaley, V. Miteva, A. Sessitch, M. A. Petrova, V. S. Soina, G. King, for providing Exiguobacterium isolates and S. Tiquia for γ-Proteobacteria isolate. We thank R. M. Siletzky for laboratory assistance in portions of the study. This study was supported by NASA Astrobiology Institute (Grant # NCC2-1274).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Matsunaga.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vishnivetskaya, T.A., Kathariou, S. & Tiedje, J.M. The Exiguobacterium genus: biodiversity and biogeography. Extremophiles 13, 541–555 (2009). https://doi.org/10.1007/s00792-009-0243-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-009-0243-5