Abstract
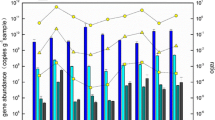
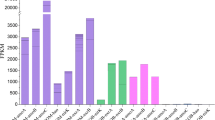
Considering their abundance and broad distribution, non-extremophilic Crenarchaeota are likely to play important roles in global organic and inorganic matter cycles. The diversity and abundance of archaeal 16S rRNA and putative ammonia monooxygenase α-subunit (amoA) genes were comparatively analyzed to study genetic potential for nitrification of ammonia-oxidizing archaea (AOA) in the surface layers (0–1 cm) of four marine sediments of the East Sea, Korea. After analysis of a 16S rRNA gene clone library, we found various archaeal groups that include the crenarchaeotal group (CG) I.1a (54.8%) and CG I.1b (5.8%), both of which are known to harbor ammonia oxidizers. Notably, the 16S rRNA gene of CG I.1b has only previously been observed in terrestrial environments. The 16S rRNA gene sequence data revealed a distinct difference in archaeal community among sites of marine sediments. Most of the obtained amoA sequences were not closely related to those of the clones retrieved from estuarine sediments and marine water columns. Furthermore, clades of unique amoA sequences were likely to cluster according to sampling sites. Using real-time PCR, quantitative analysis of amoA copy numbers showed that the copy numbers of archaeal amoA ranged from 1.1 × 107 to 4.9 × 107 per gram of sediment and were more numerous than those of bacterial amoA, with ratios ranging from 11 to 28. In conclusion, diverse CG I.1a and CG I.1b AOA inhabit surface layers of marine sediments and AOA, and especially, CG I.1a are more numerous than other ammonia-oxidizing bacteria.




Similar content being viewed by others
References
Abreu C, Jurgens G, De Marco P, Saano A, Bordalo AA (2001) Crenarchaeota and Euryarchaeota in temperate estuarine sediments. J Appl Microbiol 90:713–718
Beman JM, Francis CA (2006) Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico. Appl Environ Microbiol 72:7767–7777
Bintrim SB, Donohue TJ, Handelsman J, Roberts GP, Goodman RM (1997) Molecular phylogeny of Archaea from soil. Proc Natl Acad Sci USA 94:277–282
Buckley DH, Graber JR, Schmidt TM (1998) Phylogenetic analysis of nonthermophilic members of the kingdom crenarchaeota and their diversity and abundance in soils. Appl Environ Microbiol 64:4333–4339
DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci USA 89:5685–5689
Felsenstein J (1985) Confidence limit on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA 102:14683–14688
Fuhrman JA, McCallum K, Davis AA (1992) Novel major archaebacterial group from marine plankton. Nature 356:148–149
Good IJ (1953) The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264
Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, DeLong EF (2006) Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol 4:e95
Hershberger KL, Barns SM, Reysenbach AL, Dawson SC, Pace NR (1996) Wide diversity of Crenarchaeota. Nature 384:420
Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJ (2001) Counting the uncountable: statistical approaches to estimating microbial diversity. Appl Environ Microbiol 67:4399–4406
Inagaki F, Suzuki M, Takai K, Oida H, Sakamoto T, Aoki K, Nealson KH, Horikoshi K (2003) Microbial communities associated with geological horizons in coastal subseafloor sediments from the sea of okhotsk. Appl Environ Microbiol 69:7224–7235
Inagaki F, Tsunogai U, Suzuki M, Kosaka A, Machiyama H, Takai K, Nunoura T, Nealson KH, Horikoshi K (2004) Characterization of C1-metabolizing prokaryotic communities in methane seep habitats at the Kuroshima Knoll, southern Ryukyu Arc, by analyzing pmoA, mmoX, mxaF, mcrA, and 16S rRNA genes. Appl Environ Microbiol 70:7445–7455
Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A, Suzuki M, Takai K, Delwiche M, Colwell FS, Nealson KH, Horikoshi K, D’Hondt S, Jorgensen BB (2006) Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc Natl Acad Sci USA 103:2815–2820
Jurgens G, Lindstrom K, Saano A (1997) Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl Environ Microbiol 63:803–805
Jurgens G, Sanno A (1999) Diversity of soil Archaea in boreal forest before, and after clear-cutting and prescribed burning. FEMS Microbiol Ecol 29:205–213
Jurgens G, Glockner F, Amann R, Saano A, Montonen L, Likolammi M, Munster U (2000) Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol Ecol 34:45–56
Karner MB, DeLong EF, Karl DM (2001) Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507–510
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambrige
Knittel K, Losekann T, Boetius A, Kort R, Amann R (2005) Diversity and distribution of methanotrophic archaea at cold seeps. Appl Environ Microbiol 71:467–479
Koch M, Rudolph C, Moissl C, Huber R (2006) A cold-loving crenarchaeon is a substantial part of a novel microbial community in cold sulphidic marsh water. FEMS Microbiol Ecol 57:55–66
Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546
Kudo Y, Shibata S, Miyaki T, Aono T, Oyaizu H (1997) Peculiar archaea found in Japanese paddy soils. Biosci Biotechnol Biochem 61:917–920
Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5:150–163
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809
Li L, Kato C, Horikoshi K (1999) Microbial diversity in sediments collected from the deepest cold-seep area, the Japan Trench. Mar Biotechnol (NY) 1:391–400
MacGregor BJ, Moser DP, Alm EW, Nealson KH, Stahl DA (1997) Crenarchaeota in Lake Michigan sediment. Appl Environ Microbiol 63:1178–1181
Massana R, Murray AE, Preston CM, DeLong EF (1997) Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol 63:50–56
Nicol GW, Glover LA, Prosser JI (2003) The impact of grassland management on archaeal community structure in upland pasture rhizosphere soil. Environ Microbiol 5:152–162
Nicol GW, Tscherko D, Embley TM, Prosser JI (2005) Primary succession of soil Crenarchaeota across a receding glacier foreland. Environ Microbiol 7:337–347
Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C (2003) Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol 5:787–797
Ottesen EA, Hong JW, Quake SR, Leadbetter JR (2006) Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science 314:1464–1467
Ovreas L, Forney L, Daae FL, Torsvik V (1997) Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 63:3367–3373
Park HD, Wells GF, Bae H, Criddle CS, Francis CA (2006) Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl Environ Microbiol 72:5643–5647
Purkhold U, Pommerening-Roser A, Juretschko S, Schmid MC, Koops HP, Wagner M (2000) Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol 66:5368–5382
Reed DW, Fujita Y, Delwiche ME, Blackwelder DB, Sheridan PP, Uchida T, Colwell FS (2002) Microbial communities from methane hydrate-bearing deep marine sediments in a forearc basin. Appl Environ Microbiol 68:3759–3770
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sandaa RA, Enger O, Torsvik V (1999) Abundance and diversity of Archaea in heavy-metal-contaminated soils. Appl Environ Microbiol 65:3293–3297
Schippers A, Neretin LN (2006) Quantification of microbial communities in near-surface and deeply buried marine sediments on the Peru continental margin using real-time PCR. Environ Microbiol 8:1251–1260
Schleper C, Holben W, Klenk HP (1997) Recovery of crenarchaeotal ribosomal DNA sequences from freshwater-lake sediments. Appl Environ Microbiol 63:321–323
Schleper C, Jurgens G, Jonuscheit M (2005) Genomic studies of uncultivated archaea. Nat Rev Microbiol 3:479–488
Sliwinski MK, Goodman RM (2004) Spatial heterogeneity of crenarchaeal assemblages within mesophilic soil ecosystems as revealed by PCR-single-stranded conformation polymorphism profiling. Appl Environ Microbiol 70:1811–1820
Stepanauskas R, Sieracki ME (2007) Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. Proc Natl Acad Sci USA 104:9052–9057
Takai K, Moser DP, DeFlaun M, Onstott TC, Fredrickson JK (2001) Archaeal diversity in waters from deep South African gold mines. Appl Environ Microbiol 67:5750–5760
Teske A (2006) Microbial communities of deep marine subsurface sediments: molecular and cultivation surveys. Geomicrobiol J 23:357–368
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk HP, Schleper C (2005) Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol 7:1985–1995
Ueda T, Suga Y, Matsuguchi T (1995) Molecular phylogenetic analysis of a soil microbial community in a soybean field. Eur J Soil Sci 46:415–421
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74
Vetriani C, Jannasch HW, MacGregor BJ, Stahl DA, Reysenbach AL (1999) Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl Environ Microbiol 65:4375–4384
Whitman WB, Coleman DC, Wiebe WJ (1998) Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95:6578–6583
Wuchter C, Abbas B, Coolen MJ, Herfort L, van Bleijswijk J, Timmers P, Strous M, Teira E, Herndl GJ, Middelburg JJ, Schouten S, Sinninghe Damste JS (2006) Archaeal nitrification in the ocean. Proc Natl Acad Sci USA 103:12317–12322
Acknowledgments
This research was supported by grants PE07010 (KOPRI Project) and the 21C Frontier Microbial Genomics and Application Center Program from the Ministry of Science and Technology, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Huang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Park, SJ., Park, BJ. & Rhee, SK. Comparative analysis of archaeal 16S rRNA and amoA genes to estimate the abundance and diversity of ammonia-oxidizing archaea in marine sediments. Extremophiles 12, 605–615 (2008). https://doi.org/10.1007/s00792-008-0165-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-008-0165-7