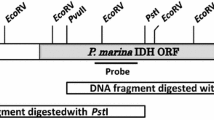
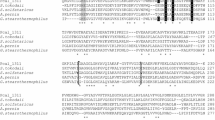
Abstract
Two structurally different monomeric and dimeric types of isocitrate dehydrogenase (IDH; EC 1.1.1.42) isozymes were confirmed to exist in a psychrophilic bacterium, Colwellia psychrerythraea, by Western blot analysis and the genes encoding them were cloned and sequenced. Open reading frames of the genes (icd-M and icd-D) encoding the monomeric and dimeric IDHs of this bacterium, IDH-M and IDH-D, were 2,232 and 1,251 bp in length and corresponded to polypeptides composed of 743 and 416 amino acids, respectively. The deduced amino acid sequences of the IDH-M and IDH-D showed high homology with those of monomeric and dimeric IDHs from other bacteria, respectively. Although the two genes were located in tandem, icd-M then icd-D, on the chromosomal DNA, a Northern blot analysis and primer extension experiment revealed that they are transcribed independent of each other. The expression of the monomeric and dimeric IDH isozyme genes in C. maris, a psychrophilic bacterium of the same genus as C. psychrerythraea, is known to be induced by low temperature and acetate, respectively, but no such induction in the expression of the C. psychrerythraea icd-M and icd-D genes was detected. IDH-M and IDH-D overexpressed in Escherichia coli were purified and characterized. In C. psychrerythraea, the IDH-M isozyme is cold-active whereas IDH-D is mesophilic, which is similar to C. maris that contains both cold-adapted and mesophilic isozymes of IDH. Experiments with chimeric enzymes between the cold-adapted monomeric IDHs of C. psychrerythraea and C. maris (IDH-M and ICD-II, respectively) suggested that the C-terminal region of the C. maris IDH-II is involved in its catalytic activity.










Similar content being viewed by others
References
Burke WF, Johanson RA, Reeves HC (1974) NADP+-specific isocitrate dehydrogenase of Escherichia coli. II. Subunit structure. Biochim Biophys Acta 351:333–340
Chen R, Yang H (2000) A highly specific monomeric isocitrate dehydrogenase from Corynebacterium glutamicum. Arch Biochem Biophys 383:238–245
Chung AE, Braginski JE (1972) Isocitrate dehydrogenase from Rhodopseudomonas spheroides: purification and characterization. Arch Biochem Biophys 153:357–367
Chung AE, Franzen JS (1969) Oxidized triphosphopyridine nucleotide specific isocitrate dehydrogenase from Azotobacter vinelandii. Isolation and characterization. Biochemistry 8:3175–3184
D’aoust JY, Kushner DJ (1972) Vibrio psychroerythrus sp. n. classification of the psychrophilic marine bacterium, NRC 1004. J Bacteriol 111:340–342
Eguchi H, Wakagi T, Oshima T (1989) A highly stable NADP-dependent isocitrate dehydrogenase from Thermus thermophilus HB8: purification and general properties. Biochim Biophys Acta 990:133–137
Eikmanns BJ, Rittmann D, Sahm H (1995) Cloning, sequence analysis, expression, and inactivation of the Corynebacterium glutamicum icd gene encoding isocitrate dehydrogenase and biochemical characterization of the enzyme. J Bacteriol 177:774–782
Fukunaga N, Yoshida S, Ishii A, Imagawa S, Takada Y, Sasaki S (1988) Isolation and characterization of a mutant defective in one of two isozymes of the isocitrate dehydrogenase from a psychrophilic bacterium, Vibrio sp. strain ABE-1. J Gen Appl Microbiol 34:457–465
Fukunaga N, Imagawa S, Sahara T, Ishii A, Suzuki M (1992) Purification and characterization of monomeric isocitrate dehydrogenase with NADP+-specificity from Vibrio parahaemolyticus Y4. J Biochem 112:849–855
Gerday C, Aittaleb M, Arpigny JL, Baise E, Chessa JP, Garsoux G, Petrescu I, Feller G (1997) Psychrophilic enzyme: a thermodynamic challenge. Biochim Biophys Acta 1342:119–131
Howard RL, Becker RR (1970) Isolation and some properties of the triphosphopyridine nucleotide isocitrate dehydrogenase from Bacillus stearothermophilus. J Biol Chem 245:3186–3194
Ishii A, Ochiai T, Imagawa S, Fukunaga N, Sasaki S, Minowa O, Mizuno Y, Shiokawa H (1987) Isozymes of isocitrate dehydrogenase from an obligately psychrophilic bacterium, Vibrio sp. strain ABE-1: purification, and modulation of activities by growth conditions. J Biochem 102:1489–1498
Ishii A, Suzuki M, Sahara T, Takada Y, Sasaki S, Fukunaga N (1993) Genes encoding two isocitrate dehydrogenase isozymes of psychrophilic bacterium, Vibrio sp. strain ABE-1. J Bacteriol 175:6873–6880
Leyland ML, Kelly DJ (1991) Purification and characterization of a monomeric isocitrate dehydrogenase with dual coenzyme specificity from the photosynthetic bacterium Phodomicribium vannielii. Eur J Biochem 202:85–93
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Mavromatis K, Tsigos I, Tzanodaskalaki M, Kokkinidis M, Bouriotis V (2002) Exploring the role of a glycine cluster in cold adaptation of an alkaline phosphatase. Eur J Biochem 269:2330–2335
Miyazaki K, Wintrode PL, Grayling RA, Rubingh DN, Arnold FH (2000) Directed evolution study of temperature adaptation in a psychrophilic enzyme. J Mol Biol 297:1015–1026
Neidhardt FC, Bloch PL, Smith DF (1974) Culture medium for enterobacteria. J Bacteriol 119:736–747
Ochiai T, Fukunaga N, Sasaki S (1979) Purification and some properties of two NADP+-specific isocitrate dehydrogenases from an obligately psychrophilic marine bacterium, Vibrio sp., strain ABE-1. J Biochem 86:377–384
Ochiai T, Fukunaga N, Sasaki S (1984) Two structurally different NADP-specific isocitrate dehydrogenases in an obligately psychrophilic bacterium, Vibrio sp. strain ABE-1. J Gen Appl Microbiol 30:479–487
Sahara T, Suzuki M, Tsuruha J, Takada Y, Fukunaga N (1999) cis-Acting elements responsible for low-temperature-inducible expression of the gene coding for the thermolabile isocitrate dehydrogenase isozyme of a psychrophilic bacterium, Vibrio sp. strain ABE-1. J Bacteriol 181:2602–2611
Sahara T, Takada Y, Takeuchi Y, Yamaoka N, Fukunaga N (2002) Cloning, sequencing, and expression of a gene encoding the monomeric isocitrate dehydrogenase of the nitrogen-fixing bacterium, Azotobacter vinelandii. Biosci Biotech Biochem 66:489–500
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Suzuki M, Sahara T, Tsuruha J, Takada Y, Fukunaga N (1995) Differential expression in Escherichia coli of the Vibrio sp. strain ABE-1 icdI and icdII genes encoding structurally different isocitrate dehydrogenase isozymes. J Bacteriol 177:2138–2142
Thorsness PE, Koshland DE Jr (1987) Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J Biol Chem 262:10422–10425
Wang Z-X, Brämer C, Steinbüchel A (2003) Two phenotypically compensating isocitrate dehydrogenases in Ralstonia eutropha. FEMS Microbiol Lett 227:9–16
Watanabe S, Takada Y, Fukunaga N (2001) Purification and characterization of a cold-adapted isocitrate lyase and a malate synthase from Colwellia maris, a psychrophilic bacterium. Biosci Biotechnol Biochem 65:1095–1103
Watanabe S, Yamaoka N, Takada Y, Fukunaga N (2002a) The cold-inducible icl gene encoding thermolabile isocitrate lyase of a psychrophilic bacterium, Colwellia maris. Microbiology 148:2579–2589
Watanabe S, Yamaoka N, Fukunaga N, Takada Y (2002b) Purification and characterization of a cold-adapted isocitrate lyase and expression analysis of the cold-inducible isocitrate lyase gene from the psychrophilic bacterium Colwellia psychrerythraea Extremophiles 6:397–405
Watanabe S, Yasutake Y, Tanaka I, Takada Y (2005) Elucidation of stability determinants of cold-adapted monomeric isocitrate dehydrogenase from a psychrophilic bacterium, Colwellia maris, by construction of chimeric enzymes. Microbiology 151:1083–1094
Yasutake Y, Watanabe S, Yao M, Takada Y, Fukunaga N, Tanaka I (2002) Structure of the monomeric isocitrate dehydrogenase: evidence of a protein monomerization by a domain duplication. Structure 10:1637–1648
Acknowledgements
We sincerely thank Dr. Isao Yumoto of the National Institute of Advanced Industrial Science and Technology for the donation of C. psychrerythraea.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Driessen
Rights and permissions
About this article
Cite this article
Maki, S., Yoneta, M. & Takada, Y. Two isocitrate dehydrogenases from a psychrophilic bacterium, Colwellia psychrerythraea. Extremophiles 10, 237–249 (2006). https://doi.org/10.1007/s00792-005-0493-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-005-0493-9