Abstract
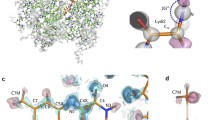
High-resolution crystal structures of Desulfovibrio vulgaris nigerythrin (DvNgr), a member of the rubrerythrin (Rbr) family, demonstrate an approximately 2-Å movement of one iron (Fe1) of the diiron site from a carboxylate to a histidine ligand upon conversion of the mixed-valent ([Fe2(II),Fe1(III)]) to diferrous states, even at cryogenic temperatures. This Glu↔His ligand “toggling” of one iron, which also occurs in DvRbr, thus, appears to be a characteristic feature of Rbr-type diiron sites. Unique features of DvNgr revealed by these structures include redox-induced flipping of a peptide carbonyl that reversibly forms a hydrogen bond to the histidine ligand to Fe1 of the diiron site, an intra-subunit proximal orientation of the rubredoxin-(Rub)-like and diiron domains, and an electron transfer pathway consisting of six covalent and two hydrogen bonds connecting the Rub-like iron with Fe2 of the diiron site. This pathway can account for DvNgr’s relatively rapid peroxidase turnover. The characteristic combination of iron sites together with the redox-dependent iron toggling between protein ligands can account for the selectivity of Rbrs for hydrogen peroxide over dioxygen.







Similar content being viewed by others
Abbreviations
- Rbr:
-
rubrerythrin
- Rub:
-
rubredoxin
- Ngr:
-
nigerythrin
- Dv:
-
Desulfovibrio vulgaris
- ENDOR:
-
Electron-nuclear double resonance
- ZnS4Rbr:
-
Rbr with zinc in the Rub-like site
- PEG:
-
polyethylene glycol
References
deMaré F, Kurtz DM Jr, Nordlund P (1996) Nat Struct Biol 3(6):539–546
Jin S et al (2002) J Am Chem Soc 124(33):9845–9855
Coulter ED et al (2000) Inorg Chim Acta 297:231–234
Sztukowska M et al (2002) Mol Microbiol 44(2):479–488
Tempel W et al (2004) Proteins Struct Funct Bioinform 57(4):878–882
Fushinobu S, Shoun H, Wakagi T (2003) Biochemistry 42(40):11707–11715
Wakagi T (2003) FEMS Microbiol Lett 222(1):33–37
LeGall J et al (1988) Biochemistry 27:1636–1642
Alban PS, Krieg NR (1998) Can J Microbiol 44:87–91
Alban PS et al (1998) J Appl Microbiol 85:875–882
Lumppio HL et al (2001) J Bacteriol 183:101–108
Coulter ED, Shenvi NV, Kurtz DM Jr (1999) Biochem Biophys Res Commun 255:317–323
Coulter ED, Kurtz DM Jr (2001) Archives Biochem Biophys 394(1):76–86
Weinberg MV et al (2004) J Bacteriol 186(23):7888–7895
Dave BC et al (1994) Biochemistry 33(12):3572–3576
Gupta N et al (1995) Biochemistry 34:3310–3318
Smoukov SK et al (2003) Biochemistry 42:6201–6208
Pierik AJ et al (1993) Eur J Biochem 212:237–245
Lumppio HL et al (1997) J Bacteriol 179:4607–4615
Bonomi F, Kurtz DM Jr, Cui X (1996) J Biol Inorg Chem 1:67–72
Sieker LC et al (2000) J Biol Inorg Chem 5(4):505–513
Li M et al (2003) J Biol Inorg Chem 80:149–155
Jin S et al (2004) J Inorg Biochem 98:786–796
Jin S et al (2004) Biochemistry 43:3204–3213
Park IY et al (2004) J Biol Inorg Chem 9(4):423–428
Eidsness MK et al (1999) Biochemistry 38(45):14803–14809
May A et al (2004) FEMS Microbiol Lett 238(1):249–254
Kennepohl P, Solomon EI (2003) Inorg Chem 42(3):696–708
Seaver LC, Imlay JA (2001) J Bacteriol 183(24):7173–7181
Jones T et al (1991) Acta Cryst A 47:110–119
Brünger AT et al (1998) Acta Crystallogr D Biol Crystallogr 54(Pt 5):905–921
Kraulis PJ (1991) J App Crystallogr 24:946–950
Gouet P, Robert X, Courcelle E (2003) Nucleic Acids Res 31(13):3320–3323
Merritt EA, Murphy MEP (1994) Acta Crystallogr Sect D Biol Crystallogr 50:869–873
Bernstein HJ (2000) Trends Biochem Sci 25(297):453–455
Acknowledgements
This work was supported by NIH grant GM40388 (D.M.K.). R.S.D. thanks the Chemistry Department of Babesh-Bolyai University, Cluj-Napoca, Romania, for a leave of absence. The authors would also like to thank B.C. Wang for providing the software ISAS and James Imlay for providing strain JI377.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article can be found at http://dx.doi.org/10.1007/s00775-005-0667-z
Ramesh B. Iyer and Radu Silaghi-Dumitrescu contributed equally to this work.
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Iyer, R.B., Silaghi-Dumitrescu, R., Kurtz, D.M. et al. High-resolution crystal structures of Desulfovibrio vulgaris (Hildenborough) nigerythrin: facile, redox-dependent iron movement, domain interface variability, and peroxidase activity in the rubrerythrins. J Biol Inorg Chem 10, 407–416 (2005). https://doi.org/10.1007/s00775-005-0650-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-005-0650-8