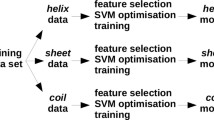
Abstract
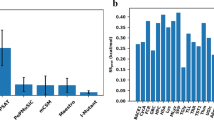
Predicting the effects of amino acid substitutions on protein stability provides invaluable information for protein design, the assignment of biological function, and for understanding disease-associated variations. To understand the effects of substitutions, computational models are preferred to time-consuming and expensive experimental methods. Several methods have been proposed for this task including machine learning-based approaches. However, models trained using limited data have performance problems and many model parameters tend to be over-fitted. To decrease the number of model parameters and to improve the generalization potential, we calculated the amino acid contact energy change for point variations using a structure-based coarse-grained model. Based on the structural properties including contact energy (CE) and further physicochemical properties of the amino acids as input features, we developed two support vector machine classifiers. M47 predicted the stability of variant proteins with an accuracy of 87 % and a Matthews correlation coefficient of 0.68 for a large dataset of 1925 variants, whereas M8 performed better when a relatively small dataset of 388 variants was used for 20-fold cross-validation. The performance of the M47 classifier on all six tested contingency table evaluation parameters is better than that of existing machine learning-based models or energy function-based protein stability classifiers.





Similar content being viewed by others
References
Bava KA, Gromiha MM, Uedaira H, Kitajima K, Sarai A (2004) ProTherm, version 4.0: thermodynamic database for proteins and mutants. Nucleic Acids Res 32(Database issue):D120–D121. doi:101093/nar/gkh08232/suppl_1/D120
Burges CJC (1998) A tutorial on support vector machines for pattern recognition. Data Min Knowl Disc 2:121–167
Capriotti E, Fariselli P, Casadio R (2004) A neural-network-based method for predicting protein stability changes upon single point mutations. Bioinformatics 20(Suppl 1):i63–i68. doi:10.1093/bioinformatics/bth92820/suppl_1/i63
Capriotti E, Fariselli P, Calabrese R, Casadio R (2005a) Predicting protein stability changes from sequences using support vector machines. Bioinformatics 21(Suppl 2):ii54–ii58. doi:10.1093/bioinformatics/bti1109
Capriotti E, Fariselli P, Casadio R (2005b) I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 33(Web Server issue):W306–W310. doi:10.1093/nar/gki375
Chang C-C, Lin C-J (2011) LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2(3):1–27. doi:10.1145/1961189.1961199
Cheng J, Randall A, Baldi P (2006) Prediction of protein stability changes for single-site mutations using support vector machines. Proteins 62(4):1125–1132. doi:10.1002/prot.20810
Collantes ER, Dunn WJ 3rd (1995) Amino acid side chain descriptors for quantitative structure-activity relationship studies of peptide analogues. J Med Chem 38(14):2705–2713
Eisenberg D, Schwarz E, Komaromy M, Wall R (1984) Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol 179(1):125–142. doi:0022-2836(84)90309-7
Fawcett T (2006) An introduction to ROC analysis. Pattern Recogn Lett 27:861–874
Ferrer-Costa C, Orozco M, de la Cruz X (2002) Characterization of disease-associated single amino acid polymorphisms in terms of sequence and structure properties. J Mol Biol 315(4):771–786. doi:20015255S0022283601952556
Gromiha MM, Uedaira H, An J, Selvaraj S, Prabakaran P, Sarai A (2002) ProTherm, thermodynamic database for proteins and mutants: developments in version 3.0. Nucleic Acids Res 30(1):301–302
Guerois R, Nielsen JE, Serrano L (2002) Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol 320(2):369–387. doi:10.1016/S0022-2836(02)00442
Kearns-Jonker M, Barteneva N, Mencel R, Hussain N, Shulkin I, Xu A, Yew M, Cramer DV (2007) Use of molecular modeling and site-directed mutagenesis to define the structural basis for the immune response to carbohydrate xenoantigens. BMC Immunol 8:3. doi:1471-2172-8-3
Keerthi SS, Lin C-J (2003) Asymptotic behaviors of support vector machines with Gaussian kernel. Neural Comput 15(7):1667–1689
Khan S, Vihinen M (2010) Performance of protein stability predictors. Hum Mutat. doi:10.1002/humu.21242
Khatun J, Khare SD, Dokholyan NV (2004) Can contact potentials reliably predict stability of proteins? J Mol Biol 336(5):1223–1238. doi:10.1016/j.jmb.2004.01
Kumar MDS, Bava KA, Gromiha MM, Prabakaran P, Kitajima K, Uedaira H, Sarai A (2006) ProTherm and ProNIT: thermodynamic databases for proteins and protein-nucleic acid interactions. Nucleic Acids Res 34:D204–D206. doi:10.1093/Nar/Gkj103
Kwasigroch JM, Gilis D, Dehouck Y, Rooman M (2002) PoPMuSiC, rationally designing point mutations in protein structures. Bioinformatics 18(12):1701–1702
Lazaridis T, Karplus M (2000) Effective energy functions for protein structure prediction. Curr Opin Struct Biol 10(2):139–145. doi:S0959-440X(00)00063-4
Lin H-T, Lin C-J (2003) A study on sigmoid kernels for SVM and the training of non-PSD kernels by SMO-type methods. National Taiwan University, Taiwan
Linden A (2006) Measuring diagnostic and predictive accuracy in disease management: an introduction to receiver operating characteristic (ROC) analysis. J Eval Clin Pract 12(2):132–139. doi:10.1111/j.1365-2753.2005.00598.x
Masso M, Vaisman II (2008) Accurate prediction of stability changes in protein mutants by combining machine learning with structure based computational mutagenesis. Bioinformatics 24(18):2002–2009. doi:10.1093/bioinformatics/btn353
Moult J (1997) Comparison of database potentials and molecular mechanics force fields. Curr Opin Struct Biol 7(2):194–199. doi:S0959-440X(97)80025-5
Rajendhran J, Gunasekaran P (2007) Molecular cloning and characterization of thermostable beta-lactam acylase with broad substrate specificity from Bacillus badius. J Biosci Bioeng 103(5):457–463. doi:10.1263/Jbb.103.457
Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L (2005) The FoldX web server: an online force field. Nucleic Acids Res 33(Web Server issue):W382–W388. doi:33/suppl_2/W382
Shen B, Vihinen M (2003) RankViaContact: ranking and visualization of amino acid contacts. Bioinformatics 19(16):2161–2162
Shen B, Bai J, Vihinen M (2008) Physicochemical feature-based classification of amino acid mutations. Protein Eng Des Sel 21(1):37–44. doi:10.1093/protein/gzm084
Thusberg J, Vihinen M (2009) Pathogenic or not? And if so, then how? Studying the effects of missense mutations using bioinformatics methods. Hum Mutat 30(5):703–714. doi:10.1002/humu.20938
Word JM, Lovell SC, LaBean TH, Taylor HC, Zalis ME, Presley BK, Richardson JS, Richardson DC (1999) Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogen atoms. J Mol Biol 285(4):1711–1733. doi:S0022-2836(98)92400-7
Yang Y, Zhou Y (2008) Specific interactions for ab initio folding of protein terminal regions with secondary structures. Proteins 72(2):793–803. doi:10.1002/prot.21968
Zamyatnin AA (1972) Protein volume in solution. Prog Biophys Mol Biol 24:107–123
Acknowledgments
This work was supported by the National Nature Science Foundation of China (31170795, 20872107), the Specialized Research Fund for the Doctoral Program of Higher Education of China (20113201110015), the Scientific Research Foundation for Returned Scholars, Ministry of Education of China, the International S&T Cooperation Program of Suzhou (SH201120) and the National 973 Programs of China (2010CB945600). The authors gratefully acknowledge the support of K·C Wong education foundation, Hong Kong, the Competitive Research Funding of Tampere University Hospital, Sigrid Juselius Foundation, and Biocenter Finland.
Conflict of interest
The authors have declared that no competing interests exist.
Author information
Authors and Affiliations
Corresponding author
Additional information
Y. Yang and B. Chen contributed equally to this work.
A website with supporting documentation and the software called PPSC (Predictor of Protein Stability Changes) is available at http://www.ibio-cn.org/softwares/PPSC/index.html and http://structure.bmc.lu.se/PPSC/.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, Y., Chen, B., Tan, G. et al. Structure-based prediction of the effects of a missense variant on protein stability. Amino Acids 44, 847–855 (2013). https://doi.org/10.1007/s00726-012-1407-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-012-1407-7