Abstract
Here, we propose the creation of the family “Yaraviridae”, a new taxon to classify a virus infecting Acanthamoeba castellanii cells. Recently, we described the discovery of a new virus infecting free-living amoebae, yaravirus, which has features that strongly differ from those of all other viruses of amoebae described to date. Yaravirus particles are about 80 nm in diameter and have a dsDNA genome of ~45 kbp containing 74 ORFs, most of which (>90%) have no homologs in current databases. Together, these data support the creation of a new species (“Yaravirus brasiliense”), a new viral genus (here proposed as “Yaravirus”), and a new viral family (here proposed as “Yaraviridae”) to classify yaravirus and other related viruses that may be described in the future. All of them are to be included into the existing realm Varidnaviria and the kingdom Bamfordvirae, due to the presence of a major capsid protein containing a double jelly-roll fold.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dozens of viruses infecting free-living amoebae have been described in different environments and parts of the world in recent years [1,2,3,4,5,6]. In addition to several other characteristics, these viruses are predominantly distinguished by the large size of their particles and a genome encoding a large number of proteins, a large proportion of which are ORFans, proteins that do not have any homologues in current protein sequence databases, [3, 6]. Recently, we described the discovery of a new virus infecting free-living amoebae, yaravirus, which has features that strongly differ from those of all other viruses infecting free-living amoebae described to date [6]. Yaravirus is the first viral isolate infecting Acanthamoeba castellanii cultures that potentially does not belong to the nucleocytoplasmic large DNA viruses (NCLDVs). This virus harbors a dsDNA genome with an exceptionally large number of ORFans (more than 90%), including novel proteins related to the construction of capsids [6]. Due to the lack of phylogenetic information, its origins are still a mystery, and similarity analysis with other viruses is unfeasible.
This virus was isolated from samples of muddy water collected from a creek near Lake Pampulha and named after an important character from the mythological stories of the Tupi indigenous tribes (Yara, the mother of waters). The novel characteristics of this virus led us to prepare and submit a formal taxonomic proposal for creation of a new species “Yaravirus brasiliense”, a new viral genus (here proposed as “Yaravirus”), and a new viral family (here proposed as “Yaraviridae”) to classify yaravirus.
Morphological properties
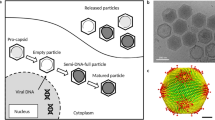
Yaravirus has a icosahedral capsid measuring about 80 nm in diameter (Fig. 1A). Cryo-electron microscopy images of purified particles suggest that yaravirus particles contain two capsid shells consisting of 26 viral proteins, one of which potentially represents a divergent capsid protein with a predicted double jelly-roll domain. This suggests that yaravirus potentially represent the first isolate of a novel group of amoebal viruses isolated from Acanthamoeba castellanii cells.
A Transmission electron micrograph of a yaravirus virion. B Yaravirus particles attached to the outside part of the host cell membrane. C Yaravirus particles undergoing endocytosis. D Yaravirus particles incorporated by the amoeba inside an endocytic vesicle. E Yaravirus particles occurring individually in the interior of endocytic vesicles. F Yaravirus particles grouped inside endocytic vesicles. G Cytoplasm of the host cell showing mitochondrial recruitment while the viral factory develops. H Mature yaravirus particles being packaged inside vesicle-like structures at a late stage of infection, suggesting exocytosis as a mechanism for the release of the virions
Prevalence and host range
The distribution and natural hosts of yaravirus remain uncertain. This virus was isolated from samples of muddy water collected from a creek near Lake Pampulha, an artificial lagoon in the city of Belo Horizonte, Brazil (19°51 0.04S and 43°58 46.00W), in June 2015. The virus was isolated in amoebas of Acanthamoeba castellanii strain Neff (ATCC 30010). After isolation, experiments were performed by infecting Acanthamoeba castellanii cells with yaravirus at a low multiplicity of infection (MOI), given the fastidious nature of this virus. Yaravirus is not able to infect and establish a productive cycle in vitro in Acanthamoeba polyphaga or Vermamoeba vermiformis.
Properties in culture
Infection with yaravirus causes a decrease in the number of viable host cells while the viral genome is replicating. During the first hours of infection, A. castellanii cultures continue to grow until about 24 hours postinfection (hpi). Replication of the viral genome then progresses slowly, and cell lysis occurs only after 72 hpi Between 96 hpi and 7 days postinfection, there is no change in the amount of yaravirus genomic DNA detected, and lysis seems to stop, with the remaining trophozoites turning into cysts. In contrast to what has been observed with other viruses infecting free-living amoebae, these results suggest participation of a host receptor in the internalization of viral particles, since, at the beginning of infection, yaravirus particles are found to be attached to the outside part of the amoebal plasma membrane (Fig. 1B–C). This is then followed by penetration of virus particles into the cell. Virions may be internalized individually or grouped inside endocytic vesicles, which, at a later stage of infection, are found next to a region previously occupied by the nucleus (Fig. 1D–F). After an eclipse phase, small crescents start to appear in the electron-lucent region of the virus factory, which is surrounded at this stage by several recruited mitochondria (Fig. 1G). Particles then develop progressively as they migrate toward the periphery of the cell. Sometimes, several yaravirus particles are packed into vesicle-like structures, suggesting that they are released by exocytosis, as is observed with other amoebal viruses (Fig. 1H). The remaining virions are released by lysis of the host cell. Although yaravirus is able to replicate in Acanthamoeba, considering its fastidious growth in this amoeba, it is possible that a different organism might act as its natural host.
Genomic and proteomic features
Yaravirus has a linear, double-stranded DNA genome, with a length of approximately 45 kbp, encoding 74 proteins. Its GC content is one of the highest among the known amoebal viruses, around 57%. Interestingly, as opposed to what has been observed for some members of the NCDLV group, the yaravirus genome does not seem to be enriched for any sequence motifs that might indicate the presence of a conserved promoter. In an intergenic region located between genes 29 and 30, there is a genomic island composed of six tRNAs (tRNA-Ser [gct], tRNA-Ser [tga], tRNA-Cys [gca], tRNA-Asn [gtt], tRNA-His [gtg], and tRNA-Ile [aat]). This is quite a remarkable number, given that a similar number of tRNAs is found in amoebal viruses with genomes much larger than that of yaravirus.
The most striking feature, however, is the number of genes with no similarity to sequences already described in databases. Genomic analysis at the nucleotide level revealed that there were no currently recognizable sequences in the yaravirus genome. The sequence similarity observed at the amino acid level is similarly low, with only six genes with distant matches (about 25–44% identity) in the nr databases. When adopting the same criteria used in genomic analysis of other amoebal viruses, 90% of the genome of the yaravirus is found to consist of sequences never described before by science. In an alternative strategy involving genome annotation based on analysis of the predicted three-dimensional structures of yaravirus proteins, the fraction of genes defined as ORFans was reduced to 80%. However, the proportion of unrecognizable proteins is still very impressive, reinforcing the idea that yaravirus is the first member of a newly discovered viral group.
These results were corroborated by proteomic analysis. The data show that yaravirus virions consist of 26 proteins. Comparisons between their predicted three-dimensional structures and structures currently available in databases have established a potential function for only five of them. Again, as observed for the genome, the proteome of yaravirus is primarily composed of proteins that have not been described previously.
Interestingly, despite representing a potential member of a newly discovered viral group, the yaraviruses seem to be exceedingly rare in nature. No sequences from the yaravirus major capsid protein (MCP) were retrieved in a search of 8,535 publicly available metagenomes generated from samples from diverse habitats across our planet. For sequences covering the yaravirus ATPase (NCVOG0249), only distant homologues in a group of endemic viruses of Pleurochrysis sp. were found, with only 33.9% identity to sequences from the same metagenomic databases.
Phylogenomics
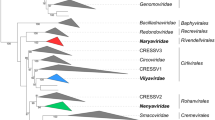
Due to the large number of ORFan genes in the yaravirus genome, most of its proteins lack the phylogenetic information necessary to incorporate this virus into an existing taxonomic group. However, as discussed above, some of its genes have sufficient similarity to genes in the current databases to be used for phylogenetic analysis. For some genes, such as gene 02 (nuclease/recombinase) and gene 69 (bifunctional DNA primase/polymerase), yaravirus groups with members of the domain Eukarya (Fig. 2A and B). For other genes, as observed for gene 46 (hypothetical protein) and gene 40 (virion packaging ATPase), yaravirus seems to be related to sequences found in giant viruses (Marseillevirus strains and members of the family Mimiviridae, respectively) (Fig. 2C and D]. A structure-guided phylogenetic analysis was also performed for hits of the yaravirus MCP protein among 235 reference genome sequences of members of the phylum Nucleocytoviricota (plus contigs of three endemic viruses of Pleurochrysis). The alignments showed an interesting, well-supported clade, grouping the yaravirus MCP with metagenomic sequences from the distant endemic viruses of Pleurochrysis sp. (Fig. 3). Further attempts were made to find other similarities between these viral groups, but only a small number of protein candidates were found, with low sequence similarity (around 24 to 33% identity). These results highlight the great differences observed between yaravirus and other amoebal viruses, supporting the creation of a new taxonomic group to include this virus.
Maximum-likelihood phylogenetic trees based on amino acid sequences of putative yaravirus proteins showing similarity to other amino acid sequences currently available in public databases. For each tree, it was constructed using amino acid sequences of yaravirus and the corresponding sequences belonging to bacteria, archaea, eukaryotes, and other members of the virosphere. Sequences were aligned using the tool ClustalW, available in MEGA X and performing 1,000 bootstrap replicates [7]. A Predicted exonuclease/recombinase. B Bifunctional DNA primase/polymerase. C A hypothetical protein. D Virion packaging ATPase. Yaravirus is highlighted in blue, and branch support is shown as coloured circles.
A structure-guided phylogenetic analysis performed on hmm search hits for the putative yaravirus MCP protein among 235 reference genome sequences (from members of the phylum Nucleocytoviricota plus contigs of three endemic viruses of Pleurochrysis) and using hidden Markov models. The alignments were made using the tool Expresso, implemented the software T-Coffee using PDB structure files. Phylogenetic trees were constructed using the software IQ-Tree (v1.6.12; ref 7) with the LG+F+R8 model based on the built-in model select feature [8] and 1,000 ultrafast bootstrap replicates [9]. Yaravirus is highlighted in blue, and branch support is shown as coloured circles.
Conclusion
Yaravirus is a novel amoebal virus whose genome is mostly composed of ORFans. Although some of its genes show similarity to sequences already available in databases, the relationships are distant. In addition, yaravirus possesses other features that distinguish it from previously isolated viruses of amoebae. Its virion is about 80 nm in diameter, and its genome, with a size of ~45 kbp, contains 74 ORFs. Together, these data support the creation of a new species for which we propose the name “Yaravirus brasiliense”, a new viral genus (here proposed as “Yaravirus”), and a new viral family (here proposed as “Yaraviridae”), in which yaravirus and other related viruses can be properly classified. The presence of a major capsid protein containing a double jelly-roll fold would warrant the inclusion of this virus in the existing realm Varidnaviria and the kingdom Bamfordvirae. A formal taxonomic proposal has been prepared and submitted to the ICTV and is currently under consideration and awaiting a ratification vote.
References
La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie JM, Raoult D (2003) A giant virus in amoebae. Science 299(5615):2033. https://doi.org/10.1126/science.1081867
Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L, Robert C, Azza S, Sun S, Rossmann MG, Suzan-Monti M, La Scola B, Koonin EV, Raoult D (2009) Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci USA 106(51):21848–21853. https://doi.org/10.1073/pnas.0911354106
Philippe N, Legendre M, Doutre G, Coute Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie JM, Abergel C (2013) Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341(6143):281–286. https://doi.org/10.1126/science.1239181
Legendre M, Bartoli J, Shmakova L, Jeudy S, Labadie K, Adrait A, Lescot M, Poirot O, Bertaux L, Bruley C, Coute Y, Rivkina E, Abergel C, Claverie JM (2014) Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc Natl Acad Sci USA 111(11):4274–4279. https://doi.org/10.1073/pnas.1320670111
Abrahao J, Silva L, Silva LS, Khalil JYB, Rodrigues R, Arantes T, Assis F, Boratto P, Andrade M, Kroon EG, Ribeiro B, Bergier I, Seligmann H, Ghigo E, Colson P, Levasseur A, Kroemer G, Raoult D, La Scola B (2018) Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat Commun 9(1):749. https://doi.org/10.1038/s41467-018-03168-1
Boratto PVM, Oliveira GP, Machado TB, Andrade A, Baudoin JP, Klose T, Schulz F, Azza S, Decloquement P, Chabriere E, Colson P, Levasseur A, La Scola B, Abrahao JS (2020) Yaravirus: a novel 80-nm virus infecting Acanthamoeba castellanii. Proc Natl Acad Sci USA 117(28):16579–16586. https://doi.org/10.1073/pnas.2001637117
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. https://doi.org/10.1038/nmeth.4285
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS (2018) UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522. https://doi.org/10.1093/molbev/msx281
Acknowledgements
We would like to thank Dr. Bernard La Scola (Aix Marseille Université) for the fruitful collaboration on topics related to amoebal viruses in the last years. We also thank colleagues from the Microscopy Center—UFMG and Laboratório de Vírus—UFMG for their technical support. We also thank FAPEMIG, CNPq, CAPES, and Pro-Reitoria de Pesquisa e Pós-Graduação-UFMG for financial support and scholarships. JSA is a CNPq researcher.
Funding
We thank FAPEMIG (Grant No. PPM-00732-18), CNPq, CAPES, and Pro-Reitoria de Pesquisa e Pós-Graduação-UFMG for financial support and scholarships (Grant No. 02081/2018-6). JSA is a CNPq researcher.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval
SISGEN numbers: A3DAB3F, A580BBD, and ABF23CC. SISBIO numbers: 34293 and 33326
Additional information
Handling Editor: Sead Sabanadzovic.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
de Miranda Boratto, P.V., Oliveira, G.P. & Abrahão, J.S. “Yaraviridae”: a proposed new family of viruses infecting Acanthamoeba castellanii. Arch Virol 167, 711–715 (2022). https://doi.org/10.1007/s00705-021-05326-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05326-1