Abstract
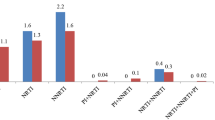
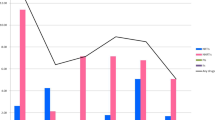
Human immunodeficiency virus (HIV) with transmitted drug-resistance (TDR) limits the therapeutic options available for treatment-naive HIV patients. This study aimed to further our understanding of the prevalence and transmission characteristics of HIV with TDR for the application of first-line antiretroviral regimens. A total of 6578 HIV-1 protease/reverse-transcriptase sequences from treatment-naive individuals in China between 2000 and 2016 were obtained from the Los Alamos HIV Sequence Database and were analyzed for TDR. Transmission networks were constructed to determine genetic relationships. The spreading routes of large TDR clusters were identified using a Bayesian phylogeographic framework. TDR mutations were detected in 274 (4.51%) individuals, with 1.40% associated with resistance to nucleoside reverse transcriptase inhibitors, 1.52% to non-nucleoside reverse transcriptase inhibitors, and 1.87% to protease inhibitors. The most frequent mutation was M46L (58, 0.89%), followed by K103N (36, 0.55%), M46I (36, 0.55%), and M184V (26, 0.40%). The prevalence of total TDR initially decreased between 2000 and 2010 (OR = 0.83, 95% CI 0.73–0.95) and then increased thereafter (OR = 1.50, 95% CI 1.13–1.97). The proportion of sequences in a cluster (clustering rate) among HIV isolates with TDR sequences was lower than that of sequences without TDR (40.5% vs. 48.8%, P = 0.023) and increased from 27.3% in 2005–2006 to 63.6% in 2015–2016 (P < 0.001). While most TDR mutations were associated with reduced relative transmission fitness, mutation M46I was associated with higher relative transmission fitness than the wild-type strain. This study identified a low-level prevalence of TDR HIV in China during the last two decades. However, the increasing TDR HIV rate since 2010, the persistent circulation of drug resistance mutations, and the expansion of self-sustaining drug resistance reservoirs may compromise the efficacy of antiretroviral therapy programs.




Similar content being viewed by others
References
Sterne JA, Hernan MA, Ledergerber B, Tilling K, Weber R, Sendi P et al (2005) Long-term effectiveness of potent antiretroviral therapy in preventing AIDS and death: a prospective cohort study. Lancet 366(9483):378–384. https://doi.org/10.1016/s0140-6736(05)67022-5
Grinsztejn B, Hosseinipour MC, Ribaudo HJ, Swindells S, Eron J, Chen YQ et al (2014) Effects of early versus delayed initiation of antiretroviral treatment on clinical outcomes of HIV-1 infection: results from the phase 3 HPTN 052 randomised controlled trial. Lancet Infect Dis 14(4):281–290. https://doi.org/10.1016/s1473-3099(13)70692-3
Coffin JM (1995) HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267(5197):483–489. https://doi.org/10.1126/science.7824947
Gupta RK, Gregson J, Parkin N, Haile-Selassie H, Tanuri A, Andrade Forero L et al (2018) HIV-1 drug resistance before initiation or re-initiation of first-line antiretroviral therapy in low-income and middle-income countries: a systematic review and meta-regression analysis. Lancet Infect Dis 18(3):346–355. https://doi.org/10.1016/s1473-3099(17)30702-8
Ncaids N (2016) Update on the AIDS/STD epidemic in China and main response in control and prevention in December, 2015. J Chin J AIDS STD 22(2):69
Luo XL, Mo LD, Su GS, Huang JP, Wu JY, Su HZ et al (2019) Incidence and types of HIV-1 drug resistance mutation among patients failing first-line antiretroviral therapy. J Pharmacol Sci 139(4):275–279. https://doi.org/10.1016/j.jphs.2018.11.016
Kan W, Teng T, Liang S, Ma Y, Tang H, Zuohela T et al (2017) Predictors of HIV virological failure and drug resistance in Chinese patients after 48 months of antiretroviral treatment, 2008–2012: a prospective cohort study. BMJ Open 7(9):e016012. https://doi.org/10.1136/bmjopen-2017-016012
Zhao S, Feng Y, Hu J, Li Y, Zuo Z, Yan J et al (2018) Prevalence of transmitted HIV drug resistance in antiretroviral treatment naive newly diagnosed individuals in China. Sci Rep 8(1):12273. https://doi.org/10.1038/s41598-018-29202-2
Su Y, Zhang F, Liu H, Smith MK, Zhu L, Wu J et al (2014) The prevalence of HIV-1 drug resistance among antiretroviral treatment naive individuals in mainland China: a meta-analysis. PLoS ONE 9(10):e110652. https://doi.org/10.1371/journal.pone.0110652
Petersen A, Cowan SA, Nielsen J, Fischer TK, Fonager J (2018) Characterisation of HIV-1 transmission clusters and drug-resistant mutations in Denmark, 2004 to 2016. Euro Surveill 23:44. https://doi.org/10.2807/1560-7917.Es.2018.23.44.1700633
Rhee SY, Clutter D, Fessel WJ, Klein D, Slome S, Pinsky BA et al (2019) Trends in the molecular epidemiology and genetic mechanisms of transmitted human immunodeficiency virus type 1 drug resistance in a large US clinic population. Clin Infect Dis 68(2):213–221. https://doi.org/10.1093/cid/ciy453
Rossetti B, Di Giambenedetto S, Torti C, Postorino MC, Punzi G, Saladini F et al (2018) Evolution of transmitted HIV-1 drug resistance and viral subtypes circulation in Italy from 2006 to 2016. HIV Med 19(9):619–628. https://doi.org/10.1111/hiv.12640
McFaul K, Lim C, Jones R, Asboe D, Pozniak A, Sonecha S et al (2014) Transmitted antiretroviral drug resistance in treatment naive HIV-infected persons in London in 2011 to 2013. J Int AIDS Soc 17(4 Suppl 3):19747. https://doi.org/10.7448/ias.17.4.19747
Katoh K, Rozewicki J, Yamada KD (2017) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20(4):1160–1166. https://doi.org/10.1093/bib/bbx108
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Yuan H, Liu Z, Wu X, Wu M, Fang Q, Tully DC et al (2019) Evolutionary characteristics and genetic transmission patterns of predominant HIV-1 subtypes among MSM in China. Int J Infect Dis 90:125–131. https://doi.org/10.1016/j.ijid.2019.10.035
Price MN, Dehal PS, Arkin AP (2010) FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 5(3):e9490. https://doi.org/10.1371/journal.pone.0009490
Ragonnet-Cronin M, Hodcroft E, Hue S, Fearnhill E, Delpech V, Brown AJL et al (2013) Automated analysis of phylogenetic clusters. BMC Bioinformatics 14:317. https://doi.org/10.1186/1471-2105-14-317
Li X, Xue Y, Lin Y, Gai J, Zhang L, Cheng H et al (2016) Evolutionary dynamics and complicated genetic transmission network patterns of HIV-1 CRF01_AE among MSM in Shanghai. China. Sci Rep 6:34729. https://doi.org/10.1038/srep34729
Paradis E, Schliep K (2019) ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35(3):526–528. https://doi.org/10.1093/bioinformatics/bty633
Epskamp S, Cramer AOJ, Waldorp LJ, Schmittmann VD, Borsboom D (2012) qgraph: network visualizations of relationships in psychometric data. J Stat Softw 48(4):1–18. https://doi.org/10.18637/jss.v048.i04
Bouckaert R, Heled J, Kuhnert D, Vaughan T, Wu CH, Xie D et al (2014) BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10(4):e1003537. https://doi.org/10.1371/journal.pcbi.1003537
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9(8):772–772. https://doi.org/10.1038/nmeth.2109
Bielejec F, Baele G, Vrancken B, Suchard MA, Rambaut A, Lemey P (2016) Sprea D3: interactive visualization of spatiotemporal history and trait evolutionary processes. Mol Biol Evol 33(8):2167–2169. https://doi.org/10.1093/molbev/msw082
Biesinger T, Kimata JT (2008) HIV-1 transmission replication fitness and disease progression. Virol (Auckl) 2008(1):49–63
Team RC (2020) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Rhee SY, Blanco JL, Jordan MR, Taylor J, Lemey P, Varghese V et al (2015) Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: an individual-patient- and sequence-level meta-analysis. PLoS Med 12(4):e1001810. https://doi.org/10.1371/journal.pmed.1001810
Zhang F, Haberer JE, Wang Y, Zhao Y, Ma Y, Zhao D et al (2007) The Chinese free antiretroviral treatment program: challenges and responses. AIDS 21(Suppl 8):S143-148. https://doi.org/10.1097/01.aids.0000304710.10036.2b
Cao YZ, Lu HZ (2005) Care of HIV-infected patients in China. Cell Res 15(11–12):883–890. https://doi.org/10.1038/sj.cr.7290363
Dou Z, Zhang F, Zhao Y, Jin C, Zhao D, Gan X et al (2015) Progress on China’ s national free antiretroviral therapy strategy in 2002–2014. Zhonghua Liu Xing Bing Xue Za Zhi 36(12):1345–1350
2015 China AIDS response progress report (2015) National Health Family Planning Commission of the People’s Republic of China, Beijing, China. Available from: http://www.commuhealtibet.org/wp-content/uploads/2016/11/CHN_narrative_report_2015.pdf
Hofstra LM, Sauvageot N, Albert J, Alexiev I, Garcia F, Struck D et al (2016) Transmission of HIV drug resistance and the predicted effect on current first-line regimens in Europe. Clin Infect Dis 62(5):655–663. https://doi.org/10.1093/cid/civ963
Tostevin A, White E, Dunn D, Croxford S, Delpech V, Williams I et al (2017) Recent trends and patterns in HIV-1 transmitted drug resistance in the United Kingdom. HIV Med 18(3):204–213. https://doi.org/10.1111/hiv.12414
Panichsillapakit T, Smith DM, Wertheim JO, Richman DD, Little SJ, Mehta SR (2016) Prevalence of transmitted HIV drug resistance among recently infected persons in San Diego, CA 1996–2013. J Acquir Immune Defic Syndr 71(2):228–236. https://doi.org/10.1097/qai.0000000000000831
Li X, Li W, Zhong P, Fang K, Zhu K, Musa TH et al (2016) Nationwide trends in molecular epidemiology of HIV-1 in China. AIDS Res Hum Retroviruses 32(9):851–859. https://doi.org/10.1089/aid.2016.0029
Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J et al (2014) HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science 345(6193):1254031. https://doi.org/10.1126/science.1254031
Young TP, Parkin NT, Stawiski E, Pilot-Matias T, Trinh R, Kempf DJ et al (2010) Prevalence, mutation patterns, and effects on protease inhibitor susceptibility of the L76V mutation in HIV-1 protease. Antimicrob Agents Chemother 54(11):4903–4906. https://doi.org/10.1128/aac.00906-10
Funding
This study was supported by the Natural Science Foundation of China (81772170) and by the National Key Research and Development Program of China (No. 2017YFC0211704).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This study did not include experiments with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Zhongjie Shi.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yuan, H., Liu, Z., Wu, X. et al. Prevalence of transmitted HIV-1 drug resistance among treatment-naive individuals in China, 2000-2016. Arch Virol 166, 2451–2460 (2021). https://doi.org/10.1007/s00705-021-05140-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05140-9