Abstract
Coronaviruses are the paradigm of emerging 21st century zoonotic viruses, triggering numerous outbreaks and a severe global health crisis. The current COVID-19 pandemic caused by SARS-CoV-2 has affected more than 51 million people across the globe as of 12 November 2020. The crown-like spikes on the surface of the virion are the unique structural feature of viruses in the family Coronaviridae. The spike (S) protein adopts distinct conformations while mediating entry of the virus into the host. This multifunctional protein mediates the entry process by recognizing its receptor on the host cell, followed by the fusion of the viral membrane with the host cell membrane. This review article focuses on the structural and functional comparison of S proteins of the human betacoronaviruses, severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Here, we review the current state of knowledge about receptor recognition, the membrane fusion mechanism, structural epitopes, and glycosylation sites of the S proteins of these viruses. We further discuss various vaccines and other therapeutics such as monoclonal antibodies, peptides, and small molecules based on the S protein of these three viruses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coronaviruses (CoVs) are a large family of viruses that were first identified in the mid-1960s. Since then, viruses of this family have posed a major health threat to animals and humans [1]. Seven human coronaviruses (hCoVs) have been identified so far, including 229E, OC43, NL63 and HKU1, which typically infect humans around the world. Zoonotic transmission due to evolutionary events has been documented for three other hCoVs, severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [2, 3]. Three fatal outbreaks of CoV infection have occurred in the last 20 years, beginning with SARS in 2002-2003, followed by MERS in 2012, and most recently, COVID-19, caused by SARS-CoV-2 (previously 2019-nCoV) [4]. In late 2019, several pneumonia cases were reported in the city of Wuhan in China; a novel type of CoV, 2019-nCoV, now officially called SARS-CoV-2, was identified as a cause of the pneumonia outbreak. The World Health Organization (WHO) declared the novel CoV outbreak a public health emergency of international concern and later proclaimed it as the COVID-19 pandemic. This pandemic has affected 220 countries worldwide, with 51,547,733 confirmed cases as of 12 November 2020 [5]. The estimated fatality rate of COVID-19 disease is 3-4%; however, variation has been observed in different geographical regions. On the other hand, in the previous CoV outbreaks, SARS infection affected 23 countries, resulting in more than 8000 cases with a fatality rate of ~11%, and MERS-CoV infected 2494 people globally with a fatality rate of nearly 35% [6, 7]. Although the fatality rate is low for SARS-CoV-2, the total number of cases is extremely high due to the ease of transmission. To date, the most severely affected regions are the Americas with 22,203,792 cases, followed by Europe, with 13,890,009 cases, and Southeast Asia, with 9,855,189 confirmed cases [5]. The transmission rate of SARS-CoV-2 is much higher than that of MERS-CoV or SARS-CoV, consequently affecting global health and economic stability of the world.
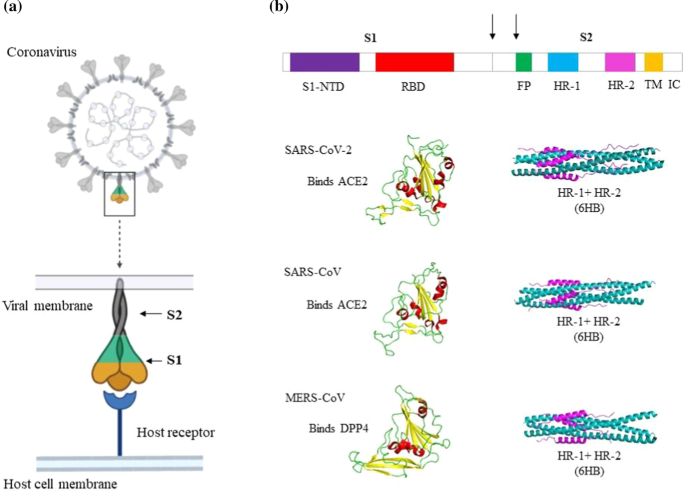
Coronaviruses belong to the family Coronaviridae and subfamily Orthocoronavirinae, which is divided into four genera, namely, Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus. SARS-CoV, MERS-CoV and SARS-CoV-2 belong to the genus Betacoronavirus, whose members infect only mammals [8]. CoVs are single-stranded RNA viruses with the largest genomes (27-32 kb) known among the RNA viruses. The genome consists of two untranslated regions (UTRs), an open reading frame (orf1a/b) encoding nonstructural proteins and other reading frames encoding the structural proteins and accessory proteins [9, 10]. Coronavirus virions are enveloped spherical particles with the spikes forming crown-like surface projections (Fig. 1a). These spike (S) proteins mediate virus entry and are responsible for determining host range. They are also the first proteins to encounter the host cell; hence, they are the primary inducer of the host immune response and are important for tissue tropism. Variations in the S proteins of diverse CoVs have allowed them to interact with a wide range of receptors and adapt to various environmental triggers for membrane fusion. In this review, we discuss the structure, function, and therapeutics of the S proteins of SARS-CoV, MERS-CoV, and SARS-CoV-2.
Schematic representation of the spike proteins of SARS-CoV-2, SARS-CoV and MERS-CoV. a) A coronavirus virion particle and its spike protein binding to the host cell receptor. b) Schematic domain organization diagram of the spike protein gene. Shown are the N-terminal domain (S1-NTD) and the receptor-binding domain (RBD) of the S1 subunit. The fusion peptide (FP), N-terminal heptad repeat (HR-1 or HR-N), and C-terminal heptad repeat (HR-2 or HR-C) of the S2 subunit are labeled. The arrows represent the two proteolysis sites. At the end of S2 subunit there is a transmembrane region (TM) and an intracellular domain (IC). The receptor-binding domain (yellow, beta sheets; red, helices; green, loops) and the S2 HR region as a 6-helix bundle (pink, HR2 or HR-C; blue, HR1 or HR-N) of SARS-CoV-2, SARS-CoV, and MERS-CoV are shown.
Receptor recognition
Receptor recognition is the first step of viral infection of the host cell. It is also a significant determinant of cross-species infection and pathogenesis. CoVs have evolved to interact with a wide variety of receptors in different hosts. CoVs belonging to different genera may bind to the same receptor, or vice-versa. SARS-CoV-2, SARS-CoV and MERS-CoV all belong to the same genus but recognize different receptors (Fig. 2). MERS-CoV recognizes dipeptidyl peptidase 4 (DPP4) as its host receptor, whereas SARS-CoV and the recently identified SARS-CoV-2 binds to angiotensin-converting enzyme 2 (ACE2) [11,12,13]. One of the two subdomains of the S protein, the S1 domain, has two distinctive units, the N-terminal (S1-NTD) domain and the C-terminal (S1-CTD) domain. Either of them can function as the receptor-binding domain (RBD). The S1-NTD is responsible for binding sugars, whereas the S1-CTD recognizes protein receptors [14,15,16,17,18]. Mouse hepatitis virus (MHV) is the sole exception, with S1-NTD binding to the protein CEACAM1 [19]. The amino acid sequence of the SARS-CoV-2 S protein is 76.3% identical to that of SARS-CoV and 29.8% identical to that of MERS-CoV. Among these three viruses, the S1 domain has more sequence diversity than the S2 domain, as the fusion core is typically conserved. The RBD consists of a core structure and a receptor-binding motif (RBM). Although SARS-like CoVs and MERS-CoV have little sequence similarity in their RBDs, the core subdomain is structurally similar in these viruses, consisting of five-stranded antiparallel beta-sheets with several short connecting alpha-helices. The RBMs, however, differ significantly, which explains why their receptor specificities differ.
Crystal structures of human betacoronavirus S1 receptor binding domains in complex with their receptor. a) Structure of the SARS-CoV-2 receptor-binding domain (RBD) showing the core domain (cyan) and the receptor-binding motif (RBM, red) complexed with human ACE2 (green; PDB ID: 6VW1). b) Structure of the SARS-CoV RBD showing the core domain (yellow) and RBM (pink) complexed with human ACE2 (green; PDB ID: 2AJF). c) Structure of the MERS-CoV RBD showing the core domain (sea green) and RBM (orange) complexed with human DPP4 (blue; PDB ID: 4L72). d) Interface between the SARS-CoV-2 RBM and ACE2. Critical residues of RBM and ACE2 involved in the interaction are labelled in black and blue, respectively. e) Interface between the SARS-CoV RBM and ACE2. Critical residues of RBM and ACE2 involved in the interaction are labelled in black and blue, respectively. f) Interface between the MERS-CoV RBM and DPP4. Interacting residues of RBM and DPP4 are labelled in black and red, respectively. The crystal structures with their respective PDB IDs were downloaded from the RCSB Protein Data Bank (https://www.rcsb.org/), and the figures were prepared using PyMol (Version 2.0 Schrodinger, LLC).
The S1 subunits of SARS-CoV-2 and SARS-CoV share 64% amino acid sequence identity [20]. Both of them interact with human ACE2 (hACE2) via their S1-CTDs, which share significant structural and sequence similarity. The crystal structures of the RBDs of these two viruses bound to the hACE2 receptor help in understanding their structural variations [21, 22]. However, the overall configuration of the RBD-ACE2 complexes of SARS-CoV and SARS-CoV-2 is identical. The RBM forms a gently concave surface that binds to the exposed outer surface of the claw-like structure of hACE2. In both of these viruses, this concave surface consists of short two-stranded antiparallel beta-sheets held on either side by two ridges formed by loops. The major structural difference between SARS-CoV and SARS-CoV-2 RBMs is the conformation of the loops in the receptor-binding ridge. SARS-CoV contains the three-residue motif Pro-Pro-Ala in this loop with a sharp turn provided by these tandem prolines. SARS-CoV-2 contains the four-residue motif Gly-Val/Gln-Glu/Thr-Gly, allowing the loop to adopt a different conformation [23]. In comparison to the RBD of SARS-CoV, the SARS-CoV-2 RBD in complex with hACE2 buries a larger surface area, and the binding interface also has more residues (17 versus 21) directly interacting with hACE2, forming more van der Waals contacts (213 versus 288) as well as H-bonds (11 versus 16) [24]. The residue Leu472 of SARS-CoV makes a weaker contact than Phe486 of SARS-CoV-2; Phe486 is inserted into the hydrophobic pocket of hACE2 and makes stronger aromatic-aromatic interactions with Tyr83. The functionally critical structural changes in the SARS-CoV-2 RBM/hACE2 interface occur near two previously detected virus-binding hotspots [25, 26]. The residues Lys31 (hotspot 31) and Lys353 (hotspot 353) of hACE2 are critical for CoV binding, as neutralization of their charge is essential for the interaction of RBM and ACE2. At SARS-CoV RBM/hACE2 interface both of these hotspot residues individually make salt bridges buried in the hydrophobic environment of the interface. At the SARS-CoV RBM/hACE2 interface, Tyr442 in the RBM supports hotspot 31, and the side chain of Tyr487 stabilizes hotspot 353. The SARS-CoV-2 RBM has evolved to stabilize these two hotspots by their rearrangement at the interface. The salt bridge formed between Lys31 and Glu35 at hotspot 31 is disrupted, and Gln493 forms hydrogen bonds individually with each of these residues. Consequently, Lys353 at the SARS-CoV-2 RBM/hACE2 interface acquires a slightly different conformation to support the hydrogen bond with the main chain of the RBM and maintain the salt bridge with Asp38 from hACE2 [23]. Outside the RBM of SARS-CoV-2, the residue Lys417 interacts and form a salt bridge with Asp30 of ACE2. In contrast, Val at the same position in SARS-CoV RBD makes no interaction with the receptor. A surface electrostatic potential comparison of the two virus-receptor interfaces revealed a positive patch contributed by Lys417 on the SARS-CoV-2 RBD, which is absent in the SARS-CoV RBD [22]. Hence, these structural features contribute to higher hACE2 binding affinity of SARS-CoV-2. However, Walls et al. reported that both SARS-CoV-2 and SARS-CoV bind to ACE2 with similar affinity [27].
The structure of the MERS-CoV S1-CTD, when compared with those of the two SARS-like CoVs, provides an interesting example of structurally similar RBDs recognizing different protein receptors [28]. Similar to the SARS-like CoVs, the RBD of MERS-CoV also has a core domain and a receptor-binding subdomain. The RBM of MERS-CoV has four-stranded antiparallel beta-sheets with a long loop connecting two of its strands, presenting a flat surface to bind to its receptor. The disulfide bond that stabilizes the receptor-binding subdomain of the RBD, is arranged differently in MERS and SARS-like viruses. The fact that its core subdomain is structurally similar to those of SARS-CoV and SARS-CoV-2 suggests that they share an evolutionary origin and that their different RBMs resulted from divergent evolution [29, 30]. The type II transmembrane protein DPP4, also called CD26, has been identified as the cellular receptor for MERS-CoV [31, 32]. DPP4 does not share any sequence or structural similarity with the receptor ACE2; it instead forms a homodimer, with each monomer containing a hydroxylase and a beta-propeller domain [33]. The MERS-CoV RBD binds laterally to the side surface of the beta-propeller domain, away from the peptidase catalytic site of DPP4 and fails to interfere with the peptidase activity of the receptor [31]. Similarly, the binding of SARS-CoV to its receptor ACE2 does not affect its enzymatic activity [34]. The binding interface of MERS-CoV/DPP4 primarily consists of a group of hydrophilic residues that form a polar contact network with hydrogen bonds and salt bridges [31]. Comparable to the hotspots at the SARS-CoV and SARS-CoV-2 receptor interface, the MERS-CoV/DPP4 interface consist of two major binding patches. In patch I, the MERS-CoV residues Glu536, Asp537 and Asp539 form a negatively charged surface, with Asp539 forming a salt bridge with Lys267 of DPP4. Also, Tyr499 in the same patch forms a hydrogen bond with the DPP4 residue Arg336. The MERS-CoV RBM forms a slightly concave outer surface accommodating a short alpha-helix of DPP4. Consequently, patch II makes a hydrophobic core consisting of Leu506, Trp553 and Val555 from MERS-CoV RBD and Leu294 and Ile295 from DPP4. A group of hydrophilic residues from both MERS-CoV RBD (Asp510, Glu513 and Tyr540) and DPP4 (His298, Arg317 and Gln344) surrounds this core. The RBM residues Asp510 and Glu513 form salt bridge and hydrogen bond interactions with DPP4 residue Arg317 and Gln344, respectively [11]. These structural studies revealed that the RBD with a conserved core domain can recognize different receptors with structural modification in the accessory subdomain. The S1 subunits of CoVs share a common evolutionary origin, but extensive divergent evolution might have resulted in their varying sequence and structure [35]. Therefore, the evolution of the receptor recognition patterns of different CoVs is a critical determinant of their host range.
Membrane fusion
S proteolysis and trigger for membrane fusion
After receptor binding, enveloped viruses rely on the fusion of their membrane with the host cell membrane. They also require a trigger mechanism, which can be low pH, proteolytic cleavage, receptor binding, or a combination thereof [36]. The characteristics of the CoV-S-protein-mediated fusion process are similar to those mediated by the class I viral fusion proteins of other viruses [37]. However, different structural features and a complex triggering mechanism that causes them to undergo conformational changes facilitating the fusion process make CoV S proteins unique. Priming of the S protein involves proteolytic cleavage at the S1/S2 interface and upstream of the fusion peptide (Fig. 3). Proteolytic processing of the S protein in most CoVs occurs later in the cell entry process, usually after receptor binding. These proteolytic cleavages mediated by the host proteases can occur at different stages of the viral life cycle. Proprotein convertases, including furin, cleave immature glycoproteins to convert them into mature ones during viral packaging. Extracellular proteases act during virus release in the extracellular space, whereas cell-surface proteases cleave after attachment of the virus to its target cell. After endocytosis of the virus particle in the target cell, lysosomal proteases such as cathepsin L and cathepsin B perform the triggering step that initiates the fusion process [38]. The requirement for these different proteases is responsible for viral tropism differences and different routes of entry. Thus, whether these viruses enter the endosome or fuse at the plasma membrane depends upon the host cell type and the availability of the required proteases. It has been reported that SARS-CoV, MERS-CoV, and SARS-CoV-2 can all be triggered to fuse at either the endosomal membrane or the plasma membrane [27, 39, 40].
Spike (S) protein models for SARS-CoV (a), MERS-CoV (b), and SARS-CoV-2 (c). Models were build using the SWISS-MODEL server to show the fusion peptide (FP) and cleavage sites (S1/S2 and S2’). The PDB IDs 6ACD, 6Q04, and 6VSB were used as templates for the SARS-CoV, MERS-CoV, and SARS-CoV-2 S model, respectively. The FP (green), S1/S2 (yellow), and S2’ (blue) cleavage sites in all of the trimers and monomers are shown. Monomers of the SARS-CoV, MERS-CoV, and SARS-CoV-2 S protein are depicted as cartoons along with sequences of the FPs and cleavage sites.
Endosomal pathway
SARS-CoV particles enter the host cell by both clathrin-dependent and clathrin-independent endocytosis pathways in the absence of exogenous proteases [41, 42]. The SARS-CoV virus S protein is not cleaved by the proprotein convertase enzyme during viral packaging [43, 44]. It therefore contains uncleaved S protein on its surface and relies on the host proteases cathepsin L and cathepsin B. This observation was confirmed when SARS-CoV infection was inhibited by either endosomal acidification inhibitors or lysosomal cysteine protease inhibitors [45, 46]. The endocytic mechanism of cell entry is a pH-dependent process, and membrane fusion occurs at low pH. However, the low pH in the endosome is not directly responsible for fusion but instead activates the lysosomal proteases that trigger fusion. The overall cell entry mechanism of MERS-CoV and SARS-CoV-2 is similar to that of SARS-CoV. MERS-CoV and SARS-CoV-2 particles can also enter the host cell through endocytosis [47, 48]. The lysosomal cysteine protease activates the MERS-CoV S protein for membrane fusion. Studies on SARS-CoV-2 have also shown that cathepsin L is essential for priming the S protein [20]. Hence, for both MERS-CoV and SARS-CoV-2, the low pH of the endosome acts as an indirect trigger for membrane fusion by activating the lysosomal protease, which in turn acts on the S protein to initiate the fusion process. In endosomal entry of SARS-CoV, cathepsin L cleaves the S protein at residue Thr678, downstream of the S1/S2 site, although cleavage site in the S2’ region remains unidentified [49]. A study reporting the potential cleavage sites in MERS-CoV suggested that cathepsin L could process the S protein at auxiliary sites [50].
Plasma membrane route
When the extracellular and cell-surface proteases are present, the virus undergoes direct fusion with the plasma membrane for immediate entry into the cell. These proteases are also involved in the activation of the CoV S protein for membrane fusion. Studies have revealed that trypsin can mediate S-protein-induced cell-cell and cell-virus fusion [40]. Trypsin treatment after receptor binding resulted in significant infection by SARS-CoV and MERS-CoV at the plasma membrane [40, 51]. Similarly, in the case of SARS-CoV-2, trypsin was found to induce cell-cell fusion by efficiently activating the S protein [20]. Different proteases, such as trypsin and thermolysin, enable the adsorption of SARS-CoV and MERS-CoV particles to the cell surface. Trypsin activates fusion of SARS-CoV by sequential cleavage at two distinct sites. The first cleavage occurs at the S1/S2 site, Arg667, which probably facilitates the second cleavage at position Arg797 near the S2’ region [52, 53]. However, the site of thermolysin cleavage remains unknown. In addition, elastase, a protease produced in the lungs during inflammation, also enhances these viral infections [54, 55]. For SARS-CoV, elastase mediates cleavage at residue Thr795, a few residues away from the fusion peptide [56]. However, for MERS-CoV and SARS-CoV-2, the exact cleavage sites of these exogenous proteases have not yet been determined. A member of the transmembrane protease, serine subfamily (TMPRSS) such as TMPRSS2 or TMPRSS4 can induce SARS-CoV and MERS-CoV fusion [57, 58]. Type II transmembrane serine proteases (TTSPs) have also been shown to affect SARS-CoV and MERS-CoV fusion. TMPRSS11a can cleave and activate the SARS-CoV S protein and trigger the fusion mechanism [59]. Studies on MERS-CoV cell fusion revealed that TMPRSS11a and TMPRSS11e could also activate the S protein [60]. Another cell membrane protease known as human airway trypsin-like protease (HAT) activates the MERS and SARS-CoV S proteins and supports viral spread in infected humans [60, 61]. TMPRSS2 is a membrane-bound serine protease that is known to activate SARS-CoV, MERS-CoV, and SARS-CoV-2 S proteins for fusion [51, 58, 62,63,64,65]. The activation site for TMPRSS2 is in the motif RSAR in the S2’ region in both MERS-CoV and SARS-CoV-2, and this step requires prior S1/S2 cleavage [50, 65], while for SARS-CoV, the TMPRSS2-mediated S protein cleavage is at Arg667, and activation near the S2’ region occurs at Arg797 [66]. Unlike SARS-CoV, the MERS-CoV S proteins are pre-cleaved by host proprotein convertases during viral packaging, as they contain a furin cleavage site (RSVR) at S1/S2. However, a two-step sequential protease cleavage model has been proposed for both SARS-CoV and MERS-CoV, involving a priming cleavage at the S1/S2 site and an activating cleavage at S2’ site [52, 67]. Hence, MERS-CoV S protein fusion occurs only when this sequential cleavage takes place, first by the furin protease in the trans-Golgi network at the S1/S2 site and, second, after virus binding to the receptor. MERS-CoV particles without the furin-cleaved S protein are unable to initiate fusion at the plasma membrane and are less infectious [54]. No prior furin cleavage is required for SARS-CoV plasma membrane fusion, although conformational changes after receptor binding or an S1/S2 cleavage event can further expose the S2’ site for membrane fusion. Recent studies have revealed unique potential furin-like cleavage at the S1/S2 region of the SARS-CoV-2 S protein [68, 69]. Hence, a MERS-CoV-like furin cleavage event at the S1/S2 site during viral packaging has been proposed for SARS-CoV-2 [27]. In conclusion, proteolysis is an essential trigger preceding CoV membrane fusion. Moreover, protease activities vary with different cell types and host species, expanding the host range of CoVs.
Mechanism of fusion core formation
The CoV S protein has been categorized as a class I fusion protein based on the structural and functional features of its fusion core [70]. As described above, the S protein, upon receptor binding, undergoes proteolytic processing and triggering to initiate the membrane fusion process. Class I fusion proteins acquire different conformations during membrane fusion: a pre-fusion native-state conformation, followed by a metastable pre-fusion conformation forming a pre-hairpin intermediate, and finally a stable post-fusion structure [71, 72]. This class of fusion proteins, including the CoV S protein, forms a homotrimer in its pre- and post-fusion conformation [73]. The fusion protein has to overcome an energy barrier in order undergo the transition from one state to another. The proteolytic processing and environmental triggers help to generate the energy for the conformational transition of CoV spikes. The S2 subunit of the S protein is an alpha-helical transmembrane protein containing a fusion peptide (FP), an N-terminal (HRN or HR1) heptad repeat, and a C-terminal (HRC or HR-2) heptad repeat, followed by a transmembrane domain (TM) and a cytoplasmic intracellular domain (IC) (Fig. 1b). The hydrophobic fusion peptide consists of a short helix and a loop, with most of the non-polar residues buried within the protein core. Initially, the S protein has a trimer conformation in its pre-fusion native state. After successful priming by a host protease at S1/S2 site, the S1 subunit dissociates, forming a pre-fusion metastable state. Subsequent fusion triggering by the required proteases allows the domains to rearrange into coiled coils of three HR1 heptad repeats, forming a thermodynamically stable pre-fusion stalk conformation (pre-hairpin intermediate). As a result, the hydrophobic fusion peptide is exposed and inserts into the target membrane. In the final stage of membrane fusion, the hairpin-intermediate refolds into a stable six-helical bundle (6HB) with the central HR1 trimeric coiled-coil onto which HR2 helices fold in an antiparallel manner to form the fusion pore [74, 75]. The post-fusion conformation appears as a dumbbell-shaped structure with the 6HB; it appears as a rod-like structure in the middle, and the region between the N-terminal end and HR-N (HR-1), as well as between HR-N and HR-C, forms a globular structure at both ends. A large amount of energy is released during this conformational transition, driving the viral and host membranes together to fuse. The overall fusion mechanism of all members of the family Coronaviridae is identical and resembles that of other class I fusion proteins. However, some distinctive features such as the long 6HB, double cleavage sites, and internal fusion peptide make them unique [76]. The structures of various CoV S protein trimers have been determined using electron microscopy [77,78,79,80]. The SARS-CoV and SARS-CoV-2 S2 subunits share 89.9% sequence identity, while the fusion core is highly conserved between MERS and SARS-like CoVs [81, 82]. The fusion core structures of SARS-CoV, MERS-and SARS-CoV-2 have been determined at atomic resolution [81,82,83]. The amino acid sequence of the HR1 domain of SARS-CoV-2 has multiple variations when compared to SARS-CoV, while the HR2 domain is identical. These changes have been reported to enhance the interaction between the HR1 and HR2 domains, which in turn increases the binding affinity and thereby enhances viral infectivity or transmissibility [82]. The viral HR1 domain is an important drug target for the development of viral fusion or entry inhibitors. Several peptide-based fusion inhibitors have been discovered for MERS and SARS CoVs [82,83,84,85].
Epitopes and glycosylation sites
The S proteins on the virion surface are the principal antigenic determinants that simulate the host immune response. There is considerable information regarding the T cell and B cell epitopes of previously emerged betacoronaviruses, such as SARS-CoV and MERS-CoV. However, various immunoinformatic and experimental studies have also revealed immunogenic regions in the SARS-CoV-2 sequence [86]. Of the viral proteins, the S protein has the most identified antigenic T cell and B cell epitopes [87]. Some of the structural epitopes of the S protein are listed in Table 1 with their PDB ID numbers. It has been observed that many T cell and B cell epitopes on the S protein are conserved between SARS-CoV and SARS-CoV-2. Since the MERS-CoV S protein shares only about ~30% sequence identity with the SARS-CoV-2 S protein, the antigenic epitopes are less likely to be conserved between these two viruses. However, a recent analysis of plasma from recovered COVID-19 patients detected IgGs that could recognize the S proteins of SARS-CoV-2, SARS-CoV, and MERS-CoV [88]. Hence, it is of utmost importance to identify the critical and conserved epitopes for design of vaccines that generate cross-protective immunity against multiple betacoronaviruses.
Glycosylation of viral envelope proteins plays a crucial role in protein folding, stability and immune evasion. Glycans often shield specific epitopes that are recognized by neutralizing antibodies and thereby facilitate immune evasion. The S protein is a single-pass type I transmembrane protein with 21 to 35 N-glycosylation sites among the different CoVs. The SARS-CoV and SARS-CoV-2 S proteins encode 22 N-linked glycosylation sites in each monomer, while S protein monomer of MERS-CoV has 23 glycan modifications (Fig. 4). Site-specific analysis of N-linked glycosylation of SARS and MERS CoV S proteins has revealed extensive heterogeneity in their glycan type [89]. The MERS-CoV S protein trimer has specific mannose clusters on the surface due to an abundance of oligomannose-type glycans. The glycans at N66, N125, N155, N166, N222, N236 and N410 on the MERS S protein are all predominantly of the oligomannose type. However, the SARS-CoV and SARS-CoV-2 S proteins do not have mannose clusters on their surface; they instead have several complex-type glycans [90]. Viruses have evolved to shield their receptor binding sites with glycans to protect themselves from neutralizing antibodies. However, the MERS-CoV receptor binding site is not obstructed by glycans, as is observed for SARS-CoV and SARS-CoV-2. These structural variations might also account for differences in the virulence and pathogenicity of these viruses.
Site-specific N-linked glycosylation of the S proteins of SARS-CoV-2, SARS-CoV and MERS-CoV. The N-linked glycan sites are represented as branches. NTD, N-terminal domain; RBD, receptor binding domain; FP, fusion peptide; HR-N, N-terminal heptad repeat; HR-C, C-terminal heptad repeat; TM, transmembrane domain
Vaccines and other therapeutics targeting the S protein
The functional significance of S protein makes it an important target for developing therapeutic agents against CoVs. It has major antigenic determinants that are responsible for inducing an immune response against the viral infection. Hence, it is a significant target for the development of vaccines and neutralizing antibodies. Various peptides and small molecules target the S protein, affecting its function and ultimately interfering with virus entry and replication. However, no anti-CoV therapeutic agents have been approved for human use. Different S-protein-based therapeutics against MERS and SARS-like CoVs are discussed in this section.
Vaccines based on the S protein
From the time of the first CoV outbreak to the latest COVID-19 pandemic, various research groups from all around the world have developed various candidate vaccines. Several S-protein-based vaccines against various CoVs have been reported, as this protein is an important target for vaccine development. There are multiple types of S-protein-based vaccines, including full-length, RBD-based and recombinant-S-protein-based vaccines, DNA/RNA vaccines, and viral-vector-based vaccines. Each of them has its own advantages and disadvantages. Several recombinant viral/bacterial vector vaccines encoding the SARS-CoV S protein are in the pre-clinical stage for SARS-CoV vaccine development. They have been shown to induce long-lasting T-cell- and B-cell-mediated immune responses [91]. Recombinant-platform vaccines employ viruses as a vector, such as parainfluenza virus, adeno-associated virus, Newcastle disease virus, and replication-defective vesicular stomatitis virus to express the S protein. These vaccines induce neutralizing antibodies and T-cell responses and decrease virus titers, eventually protecting against SARS-CoV infection [92]. Other studies have also reported the use of recombinant measles viruses, baculoviruses, and rabies virus as vectors expressing the SARS S protein to elicit an immune response in transgenic mice [93,94,95]. The use of the full-length S protein gene in a DNA vaccine also resulted in SARS CoV neutralization and immunity in mice [96]. Wang et al. identified two neutralizing regions in the SARS- CoV S protein produced from DNA vaccine plasmids encoding full-length S or parts of the S protein [97]. Furthermore, construction of DNA vaccines encoding specific regions of the S protein was used as a strategy to produce an immune response against SARS-CoV [98, 99]. The vaccine VRC-SRSDNA015-00-VP, a DNA vaccine encoding the ectodomain of the SARS S protein, has completed its phase I clinical trials [100, 101]. Another recombinant S-protein-based SARS vaccine is undergoing phase I clinical trials [102]. Various studies have reported that the RBD of the SARS-CoV S protein contains major neutralizing epitopes that react with antisera from SARS-CoV-infected mice and humans [103, 104]. Immunization of mice with the RBD subunit vaccine induces long-term protection and a cellular immune response against SARS-CoV infection [105,106,107]. Thus, the recombinant protein/peptide-based subunit vaccines containing the RBD of the S protein appear to be safe and effective [108].
The approach for the development of MERS-CoV vaccines is similar to that used for SARS-CoV vaccines. The most common viruses used as a vector for recombinant-virus-based vaccines for MERS are adenovirus and modified vaccinia virus Ankara (MVA). Various groups have carried out studies using recombinant human adenovirus type 5 (rAd5) as a vector encoding the S protein or its ectodomain, reporting successful induction of an immune response in mice [109,110,111]. However, pre-existing immunity against human adenovirus type 5 in humans has hampered its efficacy as a vector. Consequently, chimpanzee adenovirus (ChAdOx1) is used as an alternative in vector-based vaccines against MERS-CoV [112, 113]. MVA and Newcastle disease virus are also used as recombinant vectors for S-protein-based vaccines against MERS infection [114,115,116]. All of the DNA vaccines developed against MERS-CoV encode the full-length S protein or the S1 domain. Immunization of mice with DNA plasmids encoding the S1 domain elicits a strong immune response and protects mice from developing pneumonia-like clinical symptoms [117, 118]. Similar to the SARS-CoV vaccines, most of the MERS-CoV subunit vaccines have focused on the RBD of S protein. However, Jiaming et al. have shown that recombinant NTD of the S protein induces neutralizing antibodies and reduces MERS-CoV infection [119]. Currently, two viral-vector-based MERS-CoV vaccines, MERS001 and MVA-MERS-S, are undergoing phase I clinical trials [120, 121]. The DNA vaccine GLS-5300, expressing the S protein of MERS-CoV is undergoing phase I/II clinical trials [122].
The COVID-19 pandemic has prompted scientists around the world to develop a vaccine against the novel CoV. There has been remarkable progress in the development of vaccines against SARS-CoV-2 since its outbreak. Scientists are also using an immunoinformatics approach to develop peptide-based vaccine candidates [123, 124]. According to the Coalition for Epidemic Preparedness Innovations, around 115 candidate vaccines are in the R&D landscape, and among them, 73 are in the early stage of development [125]. So far, 47 vaccine candidates against SARS-CoV-2 have entered clinical trials, according to a recent report by WHO, and a few of them are listed in Table 2. Among them, Ad5-nCoV, a recombinant adenovirus type 5 vector vaccine encoding S protein, was the first one to enter phase II clinical trials [126]. Another viral-vector-based vaccine, ChAdOx1 nCoV-19, is also undergoing a phase I-II trial [127]. Other S-protein-based vaccines, BNT162 (a1, b1, b2, c2) and INO-4800, have completed phase I trials. There is a long list of candidate vaccines scheduled for phase I clinical trials in 2020 [128]. However, the success rate for a vaccine candidate to pass from pre-clinical research to phase I trials is about 41-57% [129]. Nevertheless, the earlier studies on SARS-CoV and MERS-CoV vaccine development have helped in moving SARS-CoV-2 vaccine design forward.
Antibodies targeting S protein epitopes
Structural proteins are the major antigenic determinants responsible for inducing an immune response against viral infections. As mentioned in the previous section, the S protein is the viral protein with the most antigenic epitopes for inducing T-cell and B-cell responses. Antibodies generated by B cells bind to the virus, but only a few are capable of neutralizing it. Hence, passive infusion of monoclonal antibodies (mAbs) produced against the virus are used to treat several viral infections [130]. This form of therapy is known as neutralizing-antibody-mediated protection. Various groups have developed potent mouse and human mAbs against the SARS-CoV and MERS-CoV [131,132,133]. The mouse mAbs targeting SARS-CoV S protein were shown to effectively inhibit SARS-CoV infection in human cells [134]. However, these mouse mAbs may have the potential to induce an anti-mouse antibody response in humans, giving rise to various allergic reactions. However, numerous human mAbs have shown efficacy when tested in vivo against both SARS and MERS CoV infections. The majority of mAbs for both SARS-CoV and MERS-CoV target their S protein precisely in the RBD, preventing the virus attachment. The mAbs 80R, m396, CR3014, and S230.15, produced against different strains of SARS-CoV, target epitopes in the RBD of its S protein [135,136,137]. Some mAbs against MERS-CoV targeting a non-RBD region of the S protein such as G2 and G4 show cross-reactivity and protection in transgenic mice [138]. However, there is a predominance of RBD-based mAbs for MERS-CoV, such as LCA60, MERS-4, MERS-27, m336, 4C2, and 2E6, that prevent virus-receptor interactions [139]. Two mAbs, REGN3048 and REGN3051, isolated from mice immunized with the MERS-CoV S protein are undergoing a phase I clinical trial [140]. Another MERS-CoV neutralizing antibody (nAb), SAB-301, which was isolated from transchromosomic cattle is undergoing a phase I clinical trial [141]. Current efforts in developing nAbs against SARS-CoV-2 represent initial steps towards the treatment of COVID-19. The first reported human mAbs against SARS-CoV-2 are from a Chinese research lab. Those researchers isolated two human mAbs that bind to the SARS-CoV-2 RBD, blocking its interaction with the hACE2 receptor [142]. A recently published study from Utrecht University reported a neutralizing mAb, 47D11, which targets a conserved epitope in the SARS-CoV and SARS-CoV-2 RBD and has cross-neutralizing ability without affecting receptor interactions [143]. Since SARS-CoV and SARS-CoV-2 are closely related, many researchers have investigated the cross-neutralizing ability of SARS-CoV nAbs in SARS-CoV-2 infection. However, a lengthy procedure of in vivo evaluation in animal models, pre-clinical testing, and clinical trials might cause it to take several years for a SARS-CoV-2 nAb to get approved for human use [144].
Peptides and small-molecule inhibitors
Peptide-based therapeutics have great potential to be used as antiviral drugs. The first approved antiviral peptide, enfuvirtide, is an inhibitor of the HIV fusion mechanism. This peptide is derived from HIV gp41 HR2 region and prevents the interaction between HR1 and HR2, inhibiting fusion core formation [145]. However, various peptidomimetic inhibitors have been designed by different approaches to target the entry of viruses into cells. Coronavirus S-protein-based therapeutics involve various peptides that block RBD-receptor interactions, inhibit S protein cleavage and block fusion core formation. Peptides derived from both the RBD and the virus-binding motif of ACE2 can block the interaction of S1 with ACE2, inhibiting SARS-CoV entry into the cell [146, 147]. A study has also shown that synthetic peptides corresponding to the S1-S2 cleavage site region can interfere with this cleavage and restrict the production of functional S1 and S2 subunits [148]. Multiple peptides based on the HR2 domains of S proteins from SARS-CoV and MERS-CoV have been reported. Various biological techniques have been used to show that HR2 peptides compete with the viral S protein’s HR2 domain to bind to HR1 and prevent fusion core formation, with effective concentrations in the micromolar range [85, 149, 150]. Although the HR2 region and the fusion mechanism are conserved in MERS and SARS-like CoVs, there are differences in the HR1 and HR2 binding interface that could explain the difference in their sensitivity to HR2 peptides. Also, the HR2 peptides derived from these viruses were not cross-reactive. However, a recent report describes a pan-CoV fusion inhibitor targeting the HR1 domain of various human CoVs [151]. With the emergence of SARS-CoV-2, the authors, who had previously published a study of the pan-CoV inhibitor EK1, generated various lipopeptides derived from EK1 and found EK1C4 to be the most potent fusion inhibitor of SARS-CoV-2 [82]. Another recent report from China describes an HR2-sequence-based lipopeptide fusion inhibitor (IPB02) that inhibits SARS-CoV-2 S-protein-mediated cell-cell fusion and pseudovirus transduction [152]. Various computational and experimental studies are in progress to develop a peptide-based inhibitor of the SARS-CoV-2 S protein. Regardless of their inhibitory actions, the in vivo efficacy of these peptides is essential.
Various small-molecule entry inhibitors targeting the envelope proteins of viruses have been reported in the scientific literature; however, very few are under clinical development. A few studies have reported small-molecule inhibitors of CoV S proteins blocking viral entry. After the outbreak of SARS-CoV, Kao et al. identified 104 compounds with anti-SARS-CoV activity, 18 of which targeted S protein-ACE2 mediated cell entry. Among them, VE607 had potent antiviral activity (EC50 < 10 μM) and inhibited SARS-CoV entry [153]. It has also been reported that novel small molecules based on Chinese herbal medicine can inhibit the interaction of the S protein with ACE2 and can interfere with the fusion process as well [154, 155]. A study of potential SARS-CoV entry inhibitors showed that two of them inhibited the S protein. SSAA09E2 blocked the interaction of S protein with ACE2, and SSAA09E3 prevented the fusion process [156]. The number of small molecules known to target the MERS-CoV S protein is limited. An HIV entry inhibitor, ADS-J1, targeting gp41, was found to prevent the interaction between the HR1 and HR2 of MERS-CoV, thus inhibiting MERS-CoV pseudovirus infection [157]. To identify small-molecule MERS-CoV fusion inhibitors, a study evaluated some known MERS-CoV replication inhibitors and concluded that they could also inhibit clathrin-mediated endocytosis [158]. Until now, no small molecule inhibitor against the S protein of SARS-CoV-2 has reached its pre-clinical stage. A few candidate vaccines and therapeutics undergoing clinical evaluation are listed in Table 2. Moreover, drug repurposing is being adopted as a strategy against SARS-CoV-2, and various candidate drugs are undergoing clinical trials [159,160,161]. Many studies are being carried out rapidly to develop therapeutics and combat the novel CoV.
Concluding remarks
A structural and functional comparison of the S proteins of various CoVs helps to understand the basis of their evolution and pathogenicity. A few mutations or structural changes in the spike RBD can result in virus evolution and the emergence of new strains. Hence, structure-based prediction of probable mutations in the RBD, cleavage sites, fusion peptide, and glycosylation sites of these viruses might help to predict their future evolution. Furthermore, understanding the structural basis of their receptor recognition and cell entry process may assist in elucidating cross-species infection and human-to-human transmission. Hence, these studies may enhance our understanding of the intermediate host of the novel CoV, providing greater insight into its origin. Atomic-level comparisons of S proteins may bring forth a new understanding of CoV antigenicity and aid in the development of therapeutic strategies. Structural similarities in the S protein epitopes, receptor-binding regions, and fusion core provide useful insight to develop broad-spectrum treatment against these re-emerging viruses. Various vaccines and drugs are under development to combat the ongoing pandemic caused by SARS-CoV-2. Our comparative study also provides a valuable summary for further development of COVID-19 therapeutics.
References
Coronavirus. Human coronavirus types. CDC. https://www.cdc.gov/coronavirus/types.html. Accessed 20 Apr 2020
Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF (2016) Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 24:490–502
Chakraborty C, Sharma A, Bhattacharya M, Sharma G, Lee SS (2020) The 2019 novel coronavirus disease (COVID-19) pandemic: a zoonotic prospective. Asian Pac J Trop Med 13:242–246
Yang Y, Peng F, Wang R, Guan K, Jiang T, Xu G, Sun J, Chang C (2020) The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J Autoimmun. https://doi.org/10.1016/j.jaut.2020.102434
WHO Coronavirus Disease (COVID-19) Dashboard. WHO coronavirus disease (COVID-19) dashboard. https://covid19.who.int/?gclid=CjwKCAiA17P9BRB2EiwAMvwNyBDAyVhgYzITXPTVFaMcpQSx24ZCECNoyu9Zx1CHlqzsVtrlB0TzExoC03oQAvD_BwE. Accessed 12 Nov 2020
WHO (2020) Middle East respiratory syndrome coronavirus (MERS-CoV). WHO
WHO (2012) SARS (Severe Acute Respiratory Syndrome). WHO
Gorbalenya AE, Baker SC, Baric RS et al (2020) The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544
Brian DA, Baric RS (2005) Coronavirus genome structure and replication. Curr Top Microbiol Immunol 287:1–30
Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LLM, Guan Y, Rozanov M, Spaan WJM, Gorbalenya AE (2003) Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol 331:991–1004
Wang N, Shi X, Jiang L et al (2013) Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res 23:986–993
Li W, Moore MJ, Vasllieva N et al (2003) Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454
Alejandra Tortorici M, Walls AC, Lang Y et al (2019) Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol Biol 26:481–489
Liu C, Tang J, Ma Y et al (2015) Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J Virol 89:6121–6125
Promkuntod N, van Eijndhoven REW, de Vrieze G, Gröne A, Verheije MH (2014) Mapping of the receptor-binding domain and amino acids critical for attachment in the spike protein of avian coronavirus infectious bronchitis virus. Virology 448:26–32
Godet M, Grosclaude J, Delmas B, Laude H (1994) Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J Virol 68:8008–8016
Wong SK, Li W, Moore MJ, Choe H, Farzan M (2004) A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem 279:3197–3201
Du L, Zhao G, Kou Z et al (2013) Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J Virol 87:9939–9942
Kubo H, Yamada YK, Taguchi F (1994) Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J Virol 68:5403–5410
Ou X, Liu Y, Lei X et al (2020) Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 11:1620
Li F, Li W, Farzan M, Harrison SC (2005) Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864–1868
Lan J, Ge J, Yu J et al (2020) Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. https://doi.org/10.1038/s41586-020-2180-5
Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F (2020) Structural basis of receptor recognition by SARS-CoV-2. Nature 581:221–224. https://doi.org/10.1038/s41586-020-2179-y
Wang Q, Zhang Y, Wu L et al (2020) Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. https://doi.org/10.1016/j.cell.2020.03.045
Wu K, Peng G, Wilken M, Geraghty RJ, Li F (2012) Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J Biol Chem 287:8904–8911
Wu K, Chen L, Peng G, Zhou W, Pennell CA, Mansky LM, Geraghty RJ, Li F (2011) A virus-binding hot spot on human angiotensin-converting enzyme 2 is critical for binding of two different coronaviruses. J Virol 85:5331–5337
Walls AC, Park Y-J, Tortorici MA, Wall A, Mcguire AT (2020) Correspondence DV (2020) Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 183(6):1735. https://doi.org/10.1016/j.cell.2020.11.032
Li F (2016) Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 3:237–261
Chen Y, Rajashankar KR, Yang Y, Agnihothram SS, Liu C, Lin Y-L, Baric RS, Li F (2013) Crystal structure of the receptor-binding domain from newly emerged Middle East Respiratory Syndrome coronavirus downloaded from. J Virol 87:10777–10783
Li F (2015) Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol 89:1954–1964
Lu G, Hu Y, Wang Q et al (2013) Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 500:227–231
Mou H, Raj VS, van Kuppeveld FJM, Rottier PJM, Haagmans BL, Bosch BJ (2013) The receptor binding domain of the New Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J Virol 87:9379–9383
Rasmussen HB, Branner S, Wiberg FC, Wagtmann N (2003) Crystal structure of human dipeptidyl peptidase IV/CD26 in complex with a substrate analog. Nat Struct Biol 10:19–25
Li W, Zhang C, Sui J et al (2005) Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J 24:1634–1643
Li F (2012) Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J Virology 86(5):2856–2858. https://doi.org/10.1128/JVI.06882-11
White JM, Whittaker GR (2016) Fusion of enveloped viruses in endosomes. Traffic 17:593–614
Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S (2009) The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Rev Microbiol 7:226–236
Millet JK, Whittaker GR (2015) Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res 202:120–134
Qian Z, Dominguez SR, Holmes KV (2013) Role of the spike glycoprotein of human Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS ONE 8:e76469
Simmons G, Reeves JD, Rennekamp AJ, Amberg SM, Piefer AJ, Bates P (2004) Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci USA 101:4240–4245
Wang H, Yang P, Liu K, Guo F, Zhang Y, Zhang G, Jiang C (2008) SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res 18:290–301
Inoue Y, Tanaka N, Tanaka Y, Inoue S, Morita K, Zhuang M, Hattori T, Sugamura K (2007) Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J Virol 81:8722–8729
Xiao X, Chakraborti S, Dimitrov AS, Gramatikoff K, Dimitrov DS (2003) The SARS-CoV S glycoprotein: expression and functional characterization. Biochem Biophys Res Commun 312:1159–1164
Song HC, Seo M-Y, Stadler K et al (2004) Synthesis and characterization of a native, oligomeric form of recombinant severe acute respiratory syndrome coronavirus spike glycoprotein. J Virol 78:10328–10335
Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P (2005) Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA 102:11876–11881
Huang IC, Bosch BJ, Li F et al (2006) SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J Biol Chem 281:3198–3203
Yang Y, Du L, Liu C, Wang L, Ma C, Tang J, Baric RS, Jiang S, Li F (2014) Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc Natl Acad Sci USA 111:12516–12521
Yang N, Shen HM (2020) Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int J Biol Sci 16:1724–1731
Bosch BJ, Bartelink W, Rottier PJM (2008) Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J Virol 82:8887–8890
Kleine-Weber H, Elzayat MT, Hoffmann M, Pöhlmann S (2018) Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci Rep 8:1–11
Shirato K, Kawase M, Matsuyama S (2013) Middle East Respiratory Syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J Virol 87:12552–12561
Belouzard S, Chu VC, Whittaker GR (2009) Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci USA 106:5871–5876
Watanabe R, Matsuyama S, Shirato K, Maejima M, Fukushi S, Morikawa S, Taguchi F (2008) Entry from the cell surface of severe acute respiratory syndrome coronavirus with cleaved S protein as revealed by pseudotype virus bearing cleaved S protein. J Virol 82:11985–11991
Park JE, Li K, Barlan A, Fehr AR, Perlman S, McCray PB, Gallagher T (2016) Proteolytic processing of middle east respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci USA 113:12262–12267
Matsuyama S, Ujike M, Morikawa S, Tashiro M, Taguchi F (2005) Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc Natl Acad Sci USA 102:12543–12547
Belouzard S, Madu I, Whittaker GR (2010) Elastase-mediated activation of the severe acute respiratory syndrome coronavirus spike protein at discrete sites within the S2 domain. J Biol Chem 285:22758–22763
Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T (2011) A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol 85:873–882
Glowacka I, Bertram S, Muller MA et al (2011) Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol 85:4122–4134
Kam Y-W, Okumura Y, Kido H, Ng LFP, Bruzzone R, Altmeyer R (2009) Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS ONE 4:e7870
Zmora P, Hoffmann M, Kollmus H, Moldenhauer AS, Danov O, Braun A, Winkler M, Schughart K, Stefan Pöhlmann X (2018) TMPRSS11A activates the influenza A virus hemagglutinin and the MERS coronavirus spike protein and is insensitive against blockade by HAI-1. J Biol Chem 293:13863–13873
Bertram S, Glowacka I, Muller MA et al (2011) Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J Virol 85:13363–13372
Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F (2010) Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol 84:12658–12664
Gierer S, Bertram S, Kaup F et al (2013) The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol 87:5502–5511
Matsuyama S, Nao N, Shirato K et al (2020) Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci 117:7001–7003
Hoffmann M, Kleine-Weber H, Schroeder S, Mü MA, Drosten C, Pö S (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280
Reinke LM, Spiegel M, Plegge T, Hartleib A, Nehlmeier I, Gierer S, Hoffmann M, Hofmann-Winkler H, Winkler M, Pöhlmann S (2017) Different residues in the SARS-CoV spike protein determine cleavage and activation by the host cell protease TMPRSS2. PLoS ONE 12:e0179177
Mille JK, Whittaker GR (2014) Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci USA 111:15214–15219
Jaimes JA, André NM, Chappie JS, Millet JK, Whittaker GR (2020) Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically-sensitive activation loop. J Mol Biol. https://doi.org/10.1016/j.jmb.2020.04.009
Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E (2020) The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir Res 176:104742
Bosch BJ, van der Zee R, de Haan CAM, Rottier PJM (2003) The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol 77:8801–8811
Plemper R, Hammond A (2007) Inhibition of membrane fusion as a target for antiviral therapy. Antiinfect Agents Med Chem 6:248–262
Colman PM, Lawrence MC (2003) The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol 4:309–319
Kielian M, Rey FA (2006) Virus membrane-fusion proteins: More than one way to make a hairpin. Nat Rev Microbiol 4:67–76
Walls AC, Tortorici MA, Snijder J, Xiong X, Bosch BJ, Rey FA, Veesler D (2017) Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci USA 114:11157–11162
Teissier E, Penin F, Pécheur EI (2011) Targeting cell entry of enveloped viruses as an antiviral strategy. Molecules 16:221–250
Li F, Berardi M, Li W, Farzan M, Dormitzer PR, Harrison SC (2006) Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J Virol 80:6794–6800
Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (80-) 367:1260–1263
Yuan Y, Cao D, Zhang Y et al (2017) Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun 8:15092
Gui M, Song W, Zhou H, Xu J, Chen S, Xiang Y, Wang X (2017) Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res 27:119–129
Xu Y, Liu Y, Lou Z, Qin L, Li X, Bai Z, Pang H, Tien P, Gao GF, Rao Z (2004) Structural basis for coronavirus-mediated membrane fusion: crystal structure of mouse hepatitis virus spike protein fusion core. J Biol Chem 279:30514–30522
Xu Y, Lou Z, Liu Y, Pang H, Tien P, Gao GF, Rao Z (2004) Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J Biol Chem 279:49414–49419
Xia S, Liu M, Wang C et al (2020) Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res 30:343–355
Gao J, Lu G, Qi J et al (2013) Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East Respiratory Syndrome Coronavirus. J Virol 87:13134–13140
Zhu J, Xiao G, Xu Y et al (2004) Following the rule: formation of the 6-helix bundle of the fusion core from severe acute respiratory syndrome coronavirus spike protein and identification of potent peptide inhibitors. Biochem Biophys Res Commun 319:283–288
Bosch BJ, Martina BEE, Van Der Zee R, Lepault J, Haijema BJ, Versluis C, Heck AJR, De Groot R, Osterhaus ADME, Rottier PJM (2004) Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc Natl Acad Sci USA 101:8455–8460
Bhattacharya M, Sharma AR, Mallick B, Sharma G, Lee SS, Chakraborty C (2020) Immunoinformatics approach to understand molecular interaction between multi-epitopic regions of SARS-CoV-2 spike-protein with TLR4/MD-2 complex. Infect Genet Evol 85:104587
Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A (2020) A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe 27:671-680.e2
Barnes CO, West AP, Huey- KE et al (2020) Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies ll structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell 182:1–15
Watanabe Y, Berndsen ZT, Raghwani J et al (2020) Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat Commun 11:1–10
Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M (2020) Site-specific glycan analysis of the SARS-CoV-2 spike. Science 369:330–333
Rocha CD, Caetano BC, Machado AV, Bruña-Romero O (2004) Recombinant viruses as tools to induce protective cellular immunity against infectious diseases. Int Microbiol 7:83–94
Roper RL, Rehm KE (2009) SARS vaccines: where are we? Expert Rev Vaccines 8:887–898
Liniger M, Zuniga A, Tamin A et al (2008) Induction of neutralizing antibodies and cellular immune responses against SARS coronavirus by recombinant measles viruses. Vaccine 26:2164–2174
Bai B, Lu X, Meng J, Hu Q, Mao P, Lu B, Chen Z, Yuan Z, Wang H (2008) Vaccination of mice with recombinant baculovirus expressing spike or nucleocapsid protein of SARS-like coronavirus generates humoral and cellular immune responses. Mol Immunol 45:868–875
Faber M, Lamirande EW, Roberts A, Rice AB, Koprowski H, Dietzschold B, Schnell MJ (2005) A single immunization with a rhabdovirus-based vector expressing severe acute respiratory syndrome coronavirus (SARS-CoV) S protein results in the production of high levels of SARS-CoV-neutralizing antibodies. J Gen Virol 86:1435–1440
Yang ZY, Kong WP, Huang Y, Roberts A, Murphy BR, Subbarao K, Nabel GJ (2004) A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature 428:561–564
Wang S, Chou TW, Sakhatskyy PV, Huang S, Lawrence JM, Cao H, Huang X, Lu S (2005) Identification of two neutralizing regions on the severe acute respiratory syndrome coronavirus spike glycoprotein produced from the mammalian expression system. J Virol 79:1906–1910
Wang X, Xu W, Tong D, Ni J, Gao H, Wang Y, Chu Y, Li P, Yang X, Xiong S (2008) A chimeric multi-epitope DNA vaccine elicited specific antibody response against severe acute respiratory syndrome-associated coronavirus which attenuated the virulence of SARS-CoV in vitro. Immunol Lett 119:71–77
Callendret B, Lorin V, Charneau P, Marianneau P, Contamin H, Betton JM, van der Werf S, Escriou N (2007) Heterologous viral RNA export elements improve expression of severe acute respiratory syndrome (SARS) coronavirus spike protein and protective efficacy of DNA vaccines against SARS. Virology 363:288–302
Martin JE, Louder MK, Holman LSA et al (2008) A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a Phase I clinical trial. Vaccine 26:6338–6343
Phase I study of a vaccine for severe acute respiratory syndrome (SARS)—full text view—ClinicalTrials.gov. https://www.clinicaltrials.gov/ct2/show/NCT00099463. Accessed 5 May 2020
Phase I dose escalation SARS-CoV recombinant S protein, with and without adjuvant, vaccine study—full text view—ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT01376765. Accessed 5 May 2020
He Y, Zhu Q, Liu S, Zhou Y, Yang B, Li J, Jiang S (2005) Identification of a critical neutralization determinant of severe acute respiratory syndrome (SARS)-associated coronavirus: Importance for designing SARS vaccines. Virology 334:74–82
He Y, Zhou Y, Siddiqui P, Jiang S (2004) Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry. Biochem Biophys Res Commun 325:445–452
Du L, Zhao G, He Y, Guo Y, Zheng BJ, Jiang S, Zhou Y (2007) Receptor-binding domain of SARS-CoV spike protein induces long-term protective immunity in an animal model. Vaccine 25:2832–2838
Zakhartchouk AN, Sharon C, Satkunarajah M et al (2007) Immunogenicity of a receptor-binding domain of SARS coronavirus spike protein in mice: Implications for a subunit vaccine. Vaccine 25:136–143
He Y, Zhou Y, Liu S, Kou Z, Li W, Farzan M, Jiang S (2004) Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: Implication for developing subunit vaccine. Biochem Biophys Res Commun 324:773–781
Du L, He Y, Jiang S, Zheng BJ (2008) Development of subunit vaccines against severe acute respiratory syndrome. Drugs Today 44:63–73
Hashem AM, Algaissi A, Agrawal AS et al (2019) A highly immunogenic, protective, and safe adenovirus-based vaccine expressing Middle East Respiratory Syndrome Coronavirus S1-CD40L fusion protein in a transgenic human dipeptidyl peptidase 4 mouse model. J Infect Dis 220:1558–1567
Jung SY, Kang KW, Lee EY et al (2018) Heterologous prime–boost vaccination with adenoviral vector and protein nanoparticles induces both Th1 and Th2 responses against Middle East respiratory syndrome coronavirus. Vaccine 36:3468–3476
Kim E, Okada K, Kenniston T, Raj VS, AlHajri MM, Farag EABA, AlHajri F, Osterhaus ADME, Haagmans BL, Gambotto A (2014) Immunogenicity of an adenoviral-based Middle East Respiratory Syndrome coronavirus vaccine in BALB/c mice. Vaccine 32:5975–5982
Alharbi NK, Padron-Regalado E, Thompson CP et al (2017) ChAdOx1 and MVA based vaccine candidates against MERS-CoV elicit neutralising antibodies and cellular immune responses in mice. Vaccine 35:3780–3788
Dicks MDJ, Spencer AJ, Edwards NJ, Wadell G, Bojang K, Gilbert SC, Hill AVS, Cottingham MG (2012) A novel chimpanzee adenovirus vector with low human seroprevalence: Improved systems for vector derivation and comparative immunogenicity. PLoS ONE. https://doi.org/10.1371/journal.pone.0040385
Volz A, Kupke A, Song F et al (2015) Protective efficacy of recombinant modified vaccinia virus ankara delivering Middle East Respiratory Syndrome coronavirus spike glycoprotein. J Virol 89:8651–8656
Liu R, Ge J, Wang J, Shao Y, Zhang H, Wang J, Wen Z, Bu Z (2017) Newcastle disease virus-based MERS-CoV candidate vaccine elicits high-level and lasting neutralizing antibodies in Bactrian camels. J Integr Agric 16:2264–2273
Song F, Fux R, Provacia LB, Volz A, Eickmann M, Becker S, Osterhaus ADME, Haagmans BL, Sutter G (2013) Middle East respiratory syndrome coronavirus spike protein delivered by modified vaccinia virus Ankara efficiently induces virus-neutralizing antibodies. J Virol 87:11950–11954
Muthumani K, Falzarano D, Reuschel EL et al (2015) A synthetic consensus anti-spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci Transl Med 7:301–132
Chi H, Zheng X, Wang X, Wang C, Wang H, Gai W, Perlman S, Yang S, Zhao J, Xia X (2017) DNA vaccine encoding Middle East respiratory syndrome coronavirus S1 protein induces protective immune responses in mice. Vaccine 35:2069–2075
Jiaming L, Yanfeng Y, Yao D, Yawei H, Linlin B, Baoying H, Jinghua Y, Gao GF, Chuan Q, Wenjie T (2017) The recombinant N-terminal domain of spike proteins is a potential vaccine against Middle East respiratory syndrome coronavirus (MERS-CoV) infection. Vaccine 35:10–18
Safety and immunogenicity of a candidate MERS-CoV vaccine (MERS001)—full text view—ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT03399578. Accessed 6 May 2020
Safety, tolerability and immunogenicity of vaccine candidate MVA-MERS-S—full text view—ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT03615911. Accessed 6 May 2020
Evaluate the safety, tolerability and immunogenicity study of GLS-5300 in healthy volunteers—full text view—ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT03721718. Accessed 6 May 2020
Bhattacharya M, Sharma AR, Patra P, Ghosh P, Sharma G, Patra BC, Saha RP, Lee SS, Chakraborty C (2020) A SARS-CoV-2 vaccine candidate: in-silico cloning and validation. Inform Med Unlocked 20:100394
Bhattacharya M, Sharma AR, Patra P, Ghosh P, Sharma G, Patra BC, Lee S, Chakraborty C (2020) Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): immunoinformatics approach. J Med Virol 92:618–631
Le Thanh T, Andreadakis Z, Kumar A, Gómez Román R, Tollefsen S, Saville M, Mayhew S (2020) The COVID-19 vaccine development landscape. Nat Rev Drug Discov. https://doi.org/10.1038/d41573-020-00073-5
A phase II clinical trial to evaluate the recombinant vaccine for COVID-19 (adenovirus vector)—full text view—ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT04341389. Accessed 16 June 2020
A study of a candidate COVID-19 vaccine (COV001)—full text view—ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT04324606. Accessed 16 June 2020
COVID-19 vaccine—Wikipedia. https://en.wikipedia.org/wiki/COVID-19_vaccine. Accessed 6 May 2020
Gouglas D, Le Thanh T, Henderson K, Kaloudis A, Danielsen T, Hammersland NC, Robinson JM, Heaton PM, Røttingen JA (2018) Estimating the cost of vaccine development against epidemic infectious diseases: a cost minimisation study. Lancet Glob Health 6:e1386–e1396
Graham BS, Ambrosino DM (2015) History of passive antibody administration for prevention and treatment of infectious diseases. Curr Opin HIV AIDS 10:129–134
Sui J, Li W, Murakami A, Tamin A, Matthews LJ, Wong SK, Moore MJ, Tallarico AS, Olurinde M, Choe H, Anderson LJ, Bellini WJ, Farzan M, Marasco WA (2004) Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci USA 101:2536–2541. https://doi.org/10.1073/pnas.0307140101
Corti D, Passini N, Lanzavecchia A, Zambon M (2016) Rapid generation of a human monoclonal antibody to combat Middle East respiratory syndrome. J Infect Public Health 9:231–235
Tai W, Zhao G, Sun S et al (2016) A recombinant receptor-binding domain of MERS-CoV in trimeric form protects human dipeptidyl peptidase 4 (hDPP4) transgenic mice from MERS-CoV infection. Virology 499:375–382
Lai SC, Chong PCS, Yeh CT, Liu LSJ, Jan JT, Chi HY, Liu HW, Chen A, Wang YC (2005) Characterization of neutralizing monoclonal antibodies recognizing a 15-residues epitope on the spike protein HR2 region of severe acute respiratory syndrome coronavirus (SARS-CoV). J Biomed Sci 12:711–727
Zhu Z, Chakraborti S, He Y et al (2007) Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc Natl Acad Sci USA 104:12123–12128
Greenough TC, Babcock GJ, Roberts A et al (2005) Development and characterization of a severe acute respiratory syndrome-associated coronavirus-neutralizing human monoclonal antibody that provides effective immunoprophylaxis in mice. J Infect Dis 191:507–514
Rockx B, Corti D, Donaldson E, Sheahan T, Stadler K, Lanzavecchia A, Baric R (2008) Structural basis for potent cross-neutralizing human monoclonal antibody protection against lethal human and zoonotic severe acute respiratory syndrome coronavirus challenge. J Virol 82:3220–3235
Wang L, Shi W, Joyce MG et al (2015) Evaluation of candidate vaccine approaches for MERS-CoV. Nat Commun 6:1–11
Tse LV, Meganck RM, Graham RL, Baric RS (2020) The current and future state of vaccines, antivirals and gene therapies against emerging coronaviruses. Front Microbiol 11:658
A safety, tolerability, pharmacokinetics and immunogenicity trial of co-administered MERS-CoV antibodies REGN3048 and REGN3051—full text view—ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT03301090. Accessed 6 May 2020
Beigel JH, Voell J, Kumar P, Raviprakash K, Wu H, Jiao JA, Sullivan E, Luke T, Davey RT (2018) Safety and tolerability of a novel, polyclonal human anti-MERS coronavirus antibody produced from transchromosomic cattle: a phase 1 randomised, double-blind, single-dose-escalation study. Lancet Infect Dis 18:410–418
Chen X, Li R, Pan Z et al (2020) Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell Mol Immunol 17:647–649. https://doi.org/10.1038/s41423-020-0426-7
Wang C, Li W, Drabek D, Okba NMA, van Haperen R, Osterhaus ADME, van Kuppeveld FJM, Haagmans BL, Grosveld F, Bosch B-J (2020) A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun 11:2251
Jiang S, Hillyer C, Du L (2020) Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol 41:355–359
Matthews T, Salgo M, Greenberg M, Chung J, DeMasi R, Bolognesi D (2004) Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat Rev Drug Discov 3:215–225
Han DP, Penn-Nicholson A, Cho MW (2006) Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology 350:15–25
Hu H, Li L, Kao RY et al (2005) Screening and identification of linear B-cell epitopes and entry-blocking peptide of severe acute respiratory syndrome (SARS)-associated coronavirus using synthetic overlapping peptide library. J Comb Chem 7:648–656
Zheng BJ, Guan Y, He ML et al (2005) Synthetic peptides outside the spike protein heptad repeat regions as potent inhibitors of SARS-associated coronavirus. Antivir Ther 10:393–403
Lu L, Liu Q, Zhu Y et al (2014) Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun 5:1–12
Liu S, Xiao G, Chen Y et al (2004) Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: Implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 363:938–947
Xia S, Yan L, Xu W et al (2019) A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv 5:eaav4580
Zhu Y, Yu D, Yan H, Chong H, He Y (2020) Design of potent membrane fusion inhibitors against SARS-CoV-2, an emerging coronavirus with high fusogenic activity. J Virol. https://doi.org/10.1128/JVI.00635-20
Kao RY, Tsui WHW, Lee TSW et al (2004) Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem Biol 11:1293–1299
Ho TY, Wu SL, Chen JC, Li CC, Hsiang CY (2007) Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antivir Res 74:92–101
Yi L, Li Z, Yuan K et al (2004) Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J Virol 78:11334–11339
Adedeji AO, Severson W, Jonsson C, Singh K, Weiss SR, Sarafianos SG (2013) Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J Virol 87:8017–8028
Zhao G, Du L, Ma C et al (2013) A safe and convenient pseudovirus-based inhibition assay to detect neutralizing antibodies and screen for viral entry inhibitors against the novel human coronavirus MERS-CoV. Virol J. https://doi.org/10.1186/1743-422X-10-266
Liu Q, Xia S, Sun Z, Wang Q, Du L, Lu L, Jianga S (2015) Testing of middle east respiratory syndrome coronavirus replication inhibitors for the ability to block viral entry. Antimicrob Agents Chemother 59:742–744
Scheuplein VA, Seifried J, Malczyk AH et al (2015) High secretion of interferons by human plasmacytoid dendritic cells upon recognition of middle east respiratory syndrome coronavirus. J Virol 89:3859–3869
Saha RP, Sharma AR, Singh MK, Samanta S, Bhakta S, Mandal S, Bhattacharya M, Lee SS, Chakraborty C (2020) Repurposing drugs, ongoing vaccine, and new therapeutic development initiatives against COVID-19. Front Pharmacol. https://doi.org/10.3389/fphar.2020.01258
Chakraborty C, Sharma AR, Sharma G, Bhattacharya M, Lee SS (2020) SARS-CoV-2 causing pneumonia-associated respiratory disorder (COVID-19): Diagnostic and proposed therapeutic options. Eur Rev Med Pharmacol Sci 24:4016–4026
Acknowledgements
We acknowledge the financial assistance provided by the University Grant Commission in the form of a Junior Research Fellowship (JRF) to Jyoti Verma.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Jyoti Verma declares that she has no conflict of interest. Naidu Subbarao declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by either of the authors.
Additional information
Handling Editor: Zhenhai Chen.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Verma, J., Subbarao, N. A comparative study of human betacoronavirus spike proteins: structure, function and therapeutics. Arch Virol 166, 697–714 (2021). https://doi.org/10.1007/s00705-021-04961-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-04961-y