Abstract
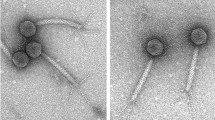
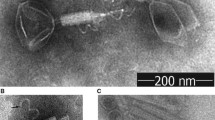
Bacillus thuringiensis (Bt) is non-pathogenic for humans and serves as a biological control agent in agriculture. Understanding its phages will help to prevent industrial production loss of Bt products and will lead to a better understanding of phages in general. The complete genome of the new B. thuringiensis phage isolate vB_BthM-Goe5 (Goe5) was sequenced, revealing a linear 157,804-bp-long dsDNA chromosome flanked by 2579-bp-long terminal repeats. It contains two tRNAs and 272 protein coding regions, 69 of which could be assigned with an annotation. Morphological investigation, using transmission electron microscopy, revealed Myoviridae morphology. The formation of a double baseplate upon tail sheath contraction indicates a link to the group of SPO1-related phages. Comparative genomics with all Bacillus-related viral genomes available in the NCBI genome database during this investigation indicated that Goe5 was a unique isolate, with Bacillus phage Bastille as its closest relative.


Similar content being viewed by others
References
Logan NA, De Vos P (2009) Systematic bacteriology. Springer, New York
Hedges SB (2002) The origin and evolution of model organisms. Nat Rev Genet 3:838–849. https://doi.org/10.1038/nrg929
Calendar R (2006) The bacteriophages. Oxford University Press, Oxford
Klumpp J, Lavigne R, Loessner MJ, Ackermann HW (2010) The SPO1-related bacteriophages. Arch Virol 155:1547–1561. https://doi.org/10.1007/s00705-010-0783-0
Hendriksen NB, Hansen BM (2006) Detection of Bacillus thuringiensis kurstaki HD1 on cabbage for human consumption. FEMS Microbiol Lett 257:106–111. https://doi.org/10.1111/j.1574-6968.2006.00159.x
Day M, Ibrahim M, Dyer D, Bulla L (2014) Genome Sequence of Bacillus thuringiensis subsp. kurstaki Strain HD-1. Genome Announc. https://doi.org/10.1128/genomeA.00613-14
Parker ML, Eiserling FA (1983) Bacteriophage SPO1 structure and morphogenesis. I. Tail structure and length regulation. J Virol 46:239–249. https://doi.org/10.1016/0923-2508(96)81062-X
Willms IM, Hoppert M, Hertel R (2017) Characterization of Bacillus subtilis Viruses vB_BsuM-Goe2 and vB_BsuM-Goe3. Viruses 9:146. https://doi.org/10.3390/v9060146
Bankevich A, Nurk S, Antipov D et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.0021
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. https://doi.org/10.1093/bioinformatics/btu153
Jones P, Binns D, Chang H-Y et al (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. https://doi.org/10.1093/bioinformatics/btu031
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci 106:19126–19131. https://doi.org/10.1073/pnas.0906412106
Kurtz S, Phillippy A, Delcher AL et al (2004) Versatile and open software for comparing large genomes. Genome Biol 5:R12. https://doi.org/10.1186/gb-2004-5-2-r12
Sauder AB, Quinn MR, Brouillette A et al (2016) Genomic characterization and comparison of seven Myoviridae bacteriophage infecting Bacillus thuringiensis. Virology 489:243–251. https://doi.org/10.1016/j.virol.2015.12.012
Gillis A, Mahillon J (2014) Phages preying on Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: past, present and future. Viruses 6:2623–2672. https://doi.org/10.3390/v6072623
Acknowledgements
We would like to thank Professor Dr. Rolf Daniel for scientific advice and guidance, and Dr. Anja Poehlein for sequencing.
Funding
This project was funded by the Volkswagen Foundation (Re. 94045).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Human and animal rights statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nordmann, B., Schilling, T., Hoppert, M. et al. Complete genome sequence of the virus isolate vB_BthM-Goe5 infecting Bacillus thuringiensis. Arch Virol 164, 1485–1488 (2019). https://doi.org/10.1007/s00705-019-04187-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04187-z