Abstract
A novel virulent bacteriophage, vB_BbrM_PHB04, infecting Bordetella bronchiseptica was isolated from wastewater collected at a swine farm in China. Phage vB_BbrM_PHB04 exhibited growth over a wide range of temperature and pH conditions and showed different efficiency of plating values and lytic spectra within the same strains at 25 °C and 37 °C. High-throughput sequencing revealed that vB_BbrM_PHB04 has a linear double-stranded DNA genome with 124 putative open reading frames. Overall, the genome of vB_BbrM_PHB04 showed very low similarity (the highest nucleotide identity 82%, 1% coverage) to other phage sequences in the GenBank database. Phylogenetic analysis indicated that vB_BbrM_PHB04 is a new member of the family Myoviridae. In addition, polymerase chain reaction-based detection of phage genes in phage-resistant B. bronchiseptica variants revealed no evidence of lysogenic activity of phage vB_BbrM_PHB04.
Similar content being viewed by others
Introduction
Bordetella bronchiseptica is a widespread Gram-negative pathogen that can be isolated from many different mammalian species. It is one of the primary causative agents of infectious respiratory diseases such as swine pneumonia and atrophic rhinitis [1]. In addition, B. bronchiseptica is often isolated in combination with other pathogens from swine suffering from pneumonia. Previous studies have shown that co-infection of viral and bacterial pathogens, including porcine reproductive and respiratory syndrome virus, porcine respiratory coronavirus, Haemophilus parasuis, Pasteurella multocida, and Streptococcus suis, exacerbates the severity of respiratory disease in pigs [2]. Importantly, human B. bronchiseptica infections have also been reported [3, 4].
B. bronchiseptica virulence factors are regulated by the complex Bordetella virulence gene product BvgAS [5]. BvgAS is a two-component signal transduction system that regulates the expression of multiple virulence factors in response to changes in environmental conditions. Different virulence factors are expressed in the three different B. bronchiseptica phases: the bvg+, bvgi, and bvg− phases. In the virulent bvg+ phase, which occurs at 37 °C in the absence of MgSO4 and nicotinic acid, B. bronchiseptica can rapidly colonize the respiratory tract of its host through the expression of adhesins, autotransporters, and toxins. The avirulent bvg− phase occurs at growth temperatures of ~25 °C or in the presence of MgSO4 or nicotinic acid. Finally, the intermediate (bvgi) phase is responsible for biofilm formation by B. bronchiseptica [5, 6].
With the increasing prevalence of multidrug-resistant bacteria, several reports have indicated the emergence of B. bronchiseptica isolates with decreased susceptibility to β-lactam antibiotics such as penicillins and cephalosporins [7]. To combat multidrug-resistant bacteria, phages offer a potential alternative to antibiotics. In the past, temperate B. bronchiseptica phages have been induced in clinical bacterial strains, but their phenotypic characteristics have rarely been described. Petrovic et al [8]. isolated 29 lytic B. bronchiseptica phages from the environment and described their phenotypic characteristics; however, the genomes of these phages were only partially sequenced to determine whether they were lytic or temperate phages. Here, we isolated a novel lytic phage from wastewater collected from a swine farm in China and studied its phenotypic and genetic characteristics. To our knowledge, the complete genomes of only three temperate B. bronchiseptica phages have been sequenced [9].
Materials and methods
Bacterial growth conditions
B. bronchiseptica and P. multocida were cultured at 37 °C for 18–30 h in tryptic soy broth (TSB; Becton, Dickinson and company, MD, USA) or on trypic soy agar (TSA; Becton, Dickinson and company, MD, USA) supplemented with 10% (v/v) sterile, defibrinated sheep blood (Jiulongbio, Zhengzhou, China). Escherichia coli was cultured at 37 °C for 12 h on TSA or TSB. All strains are available from the State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, China (Table 1).
Phage isolation
The conventional double-layer agar method was used to examine phage activity [10]. A sewage water sample (20 ml) was collected in Wuhan, China, and then sterilized by filtration through a 0.22-μm membrane. A 5-ml aliquot of the filtrate was mixed with 10 ml of an exponential phase B. bronchiseptica 4715 culture, and the mixture was incubated at 37 °C for 8 h. The culture was then centrifuged at 12,000 × g for 10 min, and the resulting supernatant was filtered using a 0.22-μm membrane. A 300-μl aliquot of the filtered supernatant was then mixed with 500 μl of B. bronchiseptica 4715, added to 6 ml of molten soft TSB (0.75% agar), supplemented with 10% sterile, defibrinated sheep blood and then poured onto the surface of a prepared TSA plate supplemented with 10% (v/v) sterile, defibrinated sheep blood. Plaques appearing after an overnight incubation at 37 °C were enumerated [10].
A single plaque was picked aseptically and re-suspended at 180 rmp in 6 ml of SM buffer (5.8 g of NaCl, 2.0 g of MgSO4·7H2O, 50 ml of Tris-HCl [pH7.4], 5.0 ml of 2% gelatin) for 3 h. The phage-containing SM buffer was collected and centrifuged at 12,000 × g for 30 s, and then the supernatant was filtered through a 0.22-μm membrane. This phage preparation was subjected to serial 10-fold dilutions in sterile SM buffer. Phage isolation by the double-layer agar method was repeated four more times, and the isolates were stored at 4 °C.
Electron microscopy
The phage filtrate was stained negatively with 2% uranyl acetate after addition of a drop of a phage suspension onto a grid surface, and then the excess stain was removed immediately. The morphology of the phages was examined under a 100-kV transmission electron microscope (HITACHI H-7650, Japan).
Thermolability and pH sensitivity
To examine these properties of the phage, we used a known method [10]. Briefly, stability of the purified phage at various temperatures (4, 20, 40, 50, 60, 70, and 80 °C) was assessed by incubation for 1 h. To evaluate the stability of the phage at different pH levels (3, 5, 7, 9, and 11), the purified phage particles were incubated at 37 °C for 1 h. Each assay was performed in triplicate, and then the samples were titered by the double-layer agar plate method.
One-step growth curve
To analyze the infectivity and replication ability of phages, a one-step growth curve test was performed as previous described [10]. Briefly, phage vB_BbrM_PHB04 was mixed with the host strain in exponential phase at a multiplicity of infection (MOI) of 0.01 and incubated at 37 °C for 5 min. The mixtures were then centrifuged at 12,000 × g for 30 s to remove any unabsorbed free phage. The precipitate was washed with 37 °C TSB, then the suspension was transferred to 30 ml of TSB supplemented with 10% (v/v) sterile, defibrinated sheep blood followed by incubation at 25 °C and 37 °C. This time point was defined as t = 0 s. At 10-min intervals, a 0.5-ml sample was collected for a total of 200 min. The titration of phage particles was conducted by the double-layer agar method. The experiment was repeated three times.
Host range determination
The bacterial strain susceptibility levels were detected using spotting methods as previously described [10]. In total, 19 B. bronchiseptica and other bacterial species (including P. multocida capsular type A, P. multocida capsular type D, E. coli O157:H7 and E. coli DH5α) were used for host range determination (Table 1). The tested bacterial strains were incubated at 25 °C and 37 °C for 16 h to allow cell lysis by the isolated phages. The efficiency of plating (EOP) values were determined by calculating the ratio of plaque-forming units (PFUs) of each phage-susceptible strain to PFUs of indicator strain (B. bronchiseptica 4715) by the double-layer agar method. This experiment was repeated three times.
DNA extraction and analysis of genome sequences
The phages’ genomic DNA was extracted using the phenol-chloroform protocol [11]. Briefly, purified phages were lysed by the addition of proteinase K (100 mg/ml), SDS (10%, wt/vol) and EDTA (0.5 mM, pH 8.0) and incubation at 56 °C in water for 2 h. The digested sample was mixed with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) centrifuged at 12,000× g (4 °C) for 10 min to remove any debris. This step was repeated three times. The supernatant was mixed with isoamyl alcohol kept at -20 °C overnight. Then the air-dried precipitate was washed three times with cold 75% ethanol. Finally, phages’ genomic DNA was dissolved in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) and was sequenced on an Illumina HiSeq 2500 sequencer with 2×100 bp read length. More than 6,200-fold coverage of phage genome was achieved by sequencing the cloned fragments. The short reads were assembled into the genome by means of SOAPdenovo (http://sourceforge.net/projects/soapdenovo2/files/SOAPdenovo2/)[12]. Protein-encoding putative open reading frames (ORFs) were predicted using Glimmer [13, 14]. The final assembled sequence was searched against the current protein and nucleotide databases (http://www.ncbi.nlm.nih.gov/) by means of the basic local alignment search tool (BLAST). Protein BLAST (BLASTP) (http://www.ncbi.nlm.nih.gov/BLAST/) was used to identify putative homologies and proteins sharing similarities with the predicted phage proteins. Phylogenetic analysis of phage large terminase subunit sequences was performed using the ClustalW program in MEGA 6 [15].
Lysogen screening
To test for possible lysogens, host B. bronchiseptica 4715 cells in exponential growth phase (approximately 107 colony-forming units/ml) were incubated with phage vB_BbrM_PHB04 at a multiplicity of infection (MOI) of 100 in SM buffer and the mixture was incubated for 10 min to allow adsorption of phages [16]. The conventional double-layer agar method was used to pick 40 random phage-resistant colonies. Single colonies were cultured with TSB supplemented with 10% (v/v) sterile, defibrinated sheep blood at 37 °C for 24 h and examined for phage vB_BbrM_PHB04 sensitivity by spot testing, applying 10 μl (107 pfu/ml) phage to visualize individual plaques. Genomic DNA was extracted from each of the bacteria resistance to phage using a commercial extraction kit (Roche, Switzerland). The PCR primers used in this study are described in Table S1. Primers were tested for specificity using the phage vB_BbrM_PHB04 genome as a positive control. This experiment was repeated three times.
Results
Morphology of phage vB_BbrM_PHB04
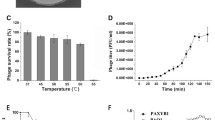
Phage vB_BbrM_PHB04 was isolated from its host, B. bronchiseptica strain 4715, which was isolated from a sewage sample from a swine farm in China. Transmission electron microscopy of the purified and concentrated phage particles showed that phage vB_BbrM_PHB04 had an isometric head (86.3 ± 1.3 nm in diameter) and a contractile tail (72.4 ± 3.2 nm long and 23.3 ± 1.8 nm wide) (Fig 1). Based on the current International Committee on Taxonomy of Viruses classification system, the phage was assigned to the family Myoviridae, order Caudovirales.
Transmission electron micrograph of phage vB_BbrM_PHB04. Phage vB_BbrM_PHB04 has an isometric polyhedral head approximately 86.3 ± 1.3 nm in diameter and a contractile tail approximately (72.4 ± 3.2 nm long and 23.3 ± 1.8 nm wide). The number of measurements is 7 (n=7) and the data are expressed as the means±SDs. Scale bar represents 100 nm. Magnification: ×150,000
Life cycle of phage vB_BbrM_PHB04
Phage vB_BbrM_PHB04 was stable at temperatures ranging from 4–50 °C, but a reduction in viral titers occurred at 60–70 °C, and titers were dramatically reduced at 80 °C (Fig 2A). The phage was stable over a broad pH range (from 5.0–9.0), but the titer dropped by approximately 1.2 log at pH 3.0 and pH 11.0 (Fig 2B). A one-step growth experiment was conducted to determine the duration of the latent period and burst size of phage vB_BbrM_PHB04. The phage had a long latent period at both 25 °C and 37 °C (~130 min and ~140 min, respectively), with an average burst size of 51 and 40 phage particles, respectively, per infected cell after 180 min (Fig 3).
One-step growth curves of phage vB_BbrM_PHB04 on Bordetella bronchiseptica 4715. The phage vB_BbrM_PHB04 had a long latent period at both 25 °C and 37 °C (~130 min and ~140 min, respectively), with an average burst size of 51 and 40 phage particles, respectively, per infected cell after 180 min. The data are expressed as the means±SDs
Host range
Nineteen B. bronchiseptica strains, including mutant B. bronchiseptica strain 3814ΔbvgA, along with four strains representing other bacterial species (including P. multocida capsular type A, P. multocida capsular type D, E. coli O157:H7 and E. coli DH5α), were included in a phage lysis assay to determine the lytic host range of phage vB_BbrM_PHB04. Seven of the B. bronchiseptica strains were sensitive to phage vB_BbrM_PHB04, while none of the other species were lysed (Table 1). For the seven sensitive B. bronchiseptica strains, the efficiency of plating (EOP) values differed slightly at different temperatures (25 °C vs. 37 °C) (Table 1). Interestingly, mutant strain 3814ΔbvgA had higher EOP values than wild-type strain 3814 (Table 1).
Genome annotation and in silico analysis
High-throughput sequencing of phage genomic DNA was performed using the Illumina HiSeq system according to the manufacturer’s instructions. The assembly of the random library of sequences yielded a closed, circular genome by means of SOAPdenovo. The complete linear, double-stranded DNA genome of phage vB_BbrM_PHB04 consisted of 94,005 bp, with a GC content of 65.5% (Fig 4). BLASTn analysis revealed that the genome of phage vB_BbrM_PHB04 showed the highest nucleotide identity (82%, though only 1% coverage) to the genome sequence of Burkholderia phage strain KACC 11889. This low sequence similarity suggested that phage vB_BbrM_PHB04 is a novel species. The genome contained 124 putative open reading frames (ORFs), and tRNA prediction, conducted using tRNAscan-SE analysis, identified three tRNA encoding genes: tRNA-Met-CAT, tRNA-Asn-GTT, and tRNA-Pro-TGG (Fig 4). The predicted ORFs encoding proteins with known functions were classified into three groups: those associated with morphogenesis and structure, those associated with DNA replication and metabolism, and those associated with lysis or lysogeny (Fig 4).
Proteins involved in structure and assembly were encoded by genes in the phage vB_BbrM_PHB04 genome. BLASTn analysis of these regions, including the baseplate (ORF3), major tail sheath (ORF12), major capsid protein (ORF20), and tail fiber protein (ORF119, ORF121, ORF122, and ORF124), showed low identity to the corresponding proteins in other phage (Fig 4; Table S1). Genes encoding DNA replication and regulation modules such as the helicase (ORF25), DNA primase (ORF98), ssDNA-binding protein (ORF101), and RecD-like DNA helicase (ORF112) were located in the middle of the phage vB_BbrM_PHB04 genome (Fig 4). However, all of these sequences showed only low levels of identity to genes encoding phage proteins of similar function. Additionally, we identified 96 genes of unknown function. With respect to lysis or lysogeny modules, ORF117 encoded a protein with 56% amino acid sequence identity to the endolysin protein of Pseudomonas phage LkO4 (Fig 4; Table S2). Holin, a protein that forms large holes in the cytoplasmic membrane of phage-infected bacteria is encoded by ORF118, showed 42% amino acid sequence identity to the holin protein from Pseudomonas phage LkO4 (Fig 4; Table S2). In addition, the protein encoded by ORF8 contained domains from multiple families, including LT_GEWL, SLT, MitE, PRK11619, and DUF3584 (Fig 4; Table S2). Proteins LT_GEWL and SLT belong to the lysozyme-like superfamily, all members of which participate in the hydrolysis of β-1,4-linked polysaccharides.
Phylogenetic analysis
The amino acid sequences of the terminase large subunit proteins (encoded by ORF55) from phage vB_BbrM_PHB04 and other phages belonging to the Myoviridae family were aligned using MEGA6 software. A neighbor-joining tree generated from these sequences revealed that phage vB_BbrM_PHB04 clustered with phages such as Bacillus phage SPO1, Bacillus phage TsarBomba, and Staphylococcus phage K in the subfamily Spounavirinae (Fig 5). This clear phylogenetic distinction from other phages and the novel characteristics of the genome indicate that phage vB_BbrM_PHB04 is a new member of the family Myoviridae, order Caudovirales.
Phylogenetic tree analysis of the phage-encoded large terminase subunit (ORF55) of phage vB_BbrM_PHB04. The amino acid sequences of the large terminase subunits were compared using MEGA6 in the analysis, and phylogenetic tree was generated using the neighbour-joining method and 1000 bootstrap replicates
Examination of lysogenic activity
The lysogenic activity of phage genes in phage-resistant bacteria was examined by PCR. In each round of screening, 40 random phage-resistant colonies were picked for further analysis and examined for vB_BbrM_PHB04 sensitivity by spot testing. The PCR results showed that the phage major capsid gene (ORF20), the terminase gene (ORF55), the helicase gene (ORF102), and the integrase gene (ORF113) were all absent from the genomes of phage-resistant colonies, suggesting that phage vB_BbrM_PHB04 does not have lysogenic activity in host bacteria (the results are not shown).
Discussion
In this study, we initially screened 17 domestic sewage samples for the presence of B. bronchiseptica phage, but failed to identify the phage in any of the samples (the results are not shown). Petrovic et al [8]. also reported a failed attempt to isolate B. bronchiseptica phage from samples collected from swine water troughs and farm wastewater. Interestingly, although we eventually isolated phage vB_BbrM_PHB04 from swine wastewater, the farm in question had no reported outbreaks or even suspected cases of bronchopneumonia or atrophic rhinitis in recent years. This may therefore indicate latent B. bronchiseptica infection of pigs.
Phage vB_BbrM_PHB04 was assigned to the family Myoviridae based on its morphological characteristics. In addition, phylogenetic analysis of the phage-encoded large terminase subunit protein revealed a close phylogenetic relationship between phage vB_BbrM_PHB04 and phages belonging to the Spounavirinae subfamily. Liu et al [9]. reported on three temperate phages isolated from B. bronchiseptica that were assigned to the family Podoviridae, while Petrovic et al [8]. isolated 29 B. bronchiseptica phages assigned to the family Siphoviridae. Based on its morphology and genome features, phage vB_BbrM_PHB04 was confirmed as a new member of the family Myoviridae, order Caudovirales.
One-step growth curve analysis revealed a significant difference in the duration of the latent period and burst size of phage vB_BbrM_PHB04 at 25 °C vs. 37 °C, which agrees with the findings of Petrovic et al [8]., who determined that the latent period and burst size of phage vB_BbrS_CN1 is dependent on temperature. In addition, the EOP values of phage vB_BbrM_PHB04 showed slight differences in phage host range at the two temperatures. B. bronchiseptica displays phenotypic variation in response to temperature change, and this phase variation may have an impact on phage receptor expression. Moreover, phage vB_BbrM_PHB04 had higher EOP values in mutant B. bronchiseptica strain 3814ΔbvgA than in wild-type strain 3814. We hypothesize that the loss of the virulence gene regulatory protein in the mutant strain resulted in enhanced vB_BbrM_PHB04 adsorption and subsequent infection.
The genome sequence of phage vB_BbrM_PHB04 showed the highest degree of nucleotide sequence identity to the genome sequence of Burkholderia phage strain KACC 11889. Functional predictions were possible for 27 of the predicted ORFs, while the remaining ORFs exhibited limited amino acid sequence identity to other hypothetical proteins, indicating that the phage vB_BbrM_PHB04 genome could be unique. In general, genome recombination occurs frequently between bacteria and phages and is conducive to phage adaptation to the host bacterium. The protein encoded by ORF113 showed 45% amino acid sequence identity to the putative integrase family protein of Pseudomonas phage PPpW-3. We know that temperate phage genomes can be integrated into bacterial genomes or plasmids and that prophages confer immunity that protects host bacteria against infection by other phages. In addition, the integrase gene is usually associated with lysogenic activity. To determine whether phage vB_BbrM_PHB04 is temperate, we designed four pairs of primers targeting essential phage genes. The results showed that none of the 40 phage-resistant bacterial clones were positive for the essential phage genes, suggesting that phage vB_BbrM_PHB04 does not have lysogenic activity.
Although no known lysogenic factors such as an excisionase, or anti-repressor proteins were identified in the genome of phage vB_BbrM_PHB04, the large number of predicted proteins with no known function may carry out lysogen-related functions. Phages containing lysogen-related genes, but not being temperate themselves, have been described previously. For example, while phage UAB_Phi20 contains all the elements needed for a lysogenic cycle, phage PPpW-3 only contains an integrase gene; however, both are lytic phages [16, 17].
We did not consider the spontaneous release of temperate phage or prophage induction from the host strain for life cycle of phage vB_BbrM_PHB04 assay. Moreover, an endolysin-encoding gene was identified in the phage vB_BbrM_PHB04 genome. Future work should therefore be conducted to express the endolysin, and to examine potential applications for this enzyme in the treatment of multidrug-resistant B. bronchiseptica.
References
Goodnow RA (1980) Biology of Bordetella bronchiseptica. Microbiol Res 44(4):722
Brockmeier SL (2004) Prior infection with Bordetella bronchiseptica increases nasal colonization by Haemophilus parasuis in swine. Vet Microbiol 99(1):75–78
Lorenzo-Pajuelo B, Villanueva JL, Rodríguez-Cuesta J, Vergara-Irigaray N, Bernabeu-Wittel M, Garcia-Curiel A, de Tejada GM (2002) Cavitary pneumonia in an AIDS patient caused by an unusual Bordetella bronchiseptica variant producing reduced amounts of pertactin and other major antigens. J Clin Microbiol 40(9):3146–3154
Yacoub AT, Katayama M, Tran J, Zadikany R, Kandula M, Greene J (2014) Bordetella bronchiseptica in the immunosuppressed population–A case series and review. Mediterr J Hematol Infect Dis 6(1):e2014031. https://doi.org/10.4084/MJHID.2014.031
Akerley BJ, Monack DM, Falkow S, Miller JF (1992) The bvgAS locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica. J Bacteriol 174(3):980–990
Vergara-Irigaray N, Chávarri-Martínez A, Rodríguez-Cuesta J, Miller JF, Cotter PA, Martínez de Tejada G (2005) Evaluation of the role of the Bvg intermediate phase in Bordetella pertussis during experimental respiratory infection. Infect Immun 73(2):748–760
Prüller S, Rensch U, Meemken D, Kaspar H, Kopp PA, Klein G, Kehrenberg C (2015) Antimicrobial susceptibility of Bordetella bronchiseptica isolates from swine and companion animals and detection of resistance genes. PloS One 10(8):e0135703. https://doi.org/10.1371/journal.pone.0135703
Petrovic A, Rakhely G, Knezevic P (2017) The first Siphoviridae family bacteriophages infecting Bordetella bronchiseptica isolated from environment. Microbial Ecol 73(2):368–377. https://doi.org/10.1007/s00248-016-0847-0
Liu M, Deora R, Doulatov SR, Gingery M, Eiserling FA, Preston A, Maskell DJ, Simons RW, Cotter PA, Parkhill J, Miller JF (2002) Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 295(5562):2091–2094
Chen YB, Sun EC, Song JY, Yang L, Wu B (2017) Complete genome sequence of a novel T7-like bacteriophage from a Pasteurella multocida capsular type A isolate. Curr Microbiol 75(5):574–579. https://doi.org/10.1007/s00284-017-1419-3
Yang Y, Cai L, Ma R, Xu Y, Tong Y, Huang Y, Jiao N, Zhang R (2017) A novel roseosiphophage isolated from the oligotrophic South China Sea. Viruses 9(5):109. https://doi.org/10.3390/v9050109
Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, Li S, Yang H, Wang J, Wang J (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272. https://doi.org/10.1101/gr.097261.109
Salzberg SL, Delcher AL, Kasif S, White O (1998) Microbial gene identification using interpolated Markov models. Nucleic Acids Res 26:544–548
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Kawato Y, Yasuike M, Nakamura Y, Shigenobu Y, Fujiwara A, Sano M, Nakai T (2014) Complete genome sequence analysis of two Pseudomonas plecoglossicida phages, potential therapeutic agents. Appl Environmen Microbiol 81(3):874–881. https://doi.org/10.1128/AEM.03038-14
Bardina C, Colom J, Spricigo DA, Otero J, Sánchez-Osuna M, Cortés P, Llagostera M (2016) Genomics of three new bacteriophages useful in the biocontrol of Salmonella. Front Microbiol 20(7):545. https://doi.org/10.3389/fmicb.2016.00545
Acknowledgements
The complete genome sequence of phage vB_BbrM_PHB04 with annotation has been deposited in GenBank under accession number MF663786.
Funding
This study was funded by Research projects of agricultural public welfare industry of China (201403054), and National Science supported planning projects of Hubei Province of China (2014BBB010).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors. I would like to declare on behalf of my co-authors that we have no conflict of interest.
Additional information
Handling Editor: Tim Skern.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, Y., Yang, L., Sun, E. et al. Characterisation of a newly detected bacteriophage infecting Bordetella bronchiseptica in swine. Arch Virol 164, 33–40 (2019). https://doi.org/10.1007/s00705-018-4034-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-4034-0