Abstract
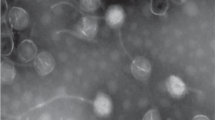
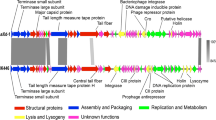
A virulent halovirus QHHSV-1 which lyses Halomonas ventosae QH52-2 originating from the Qiaohou salt mine in Yunnan, Southwest China was characterized. The complete genome of QHHSV-1 is composed of a circular double-stranded DNA of 37,270 base pairs in length, with 66.8% G+C content and 69 putative open reading frames (ORFs), which were classified into five functional groups, including morphogenesis, replication/regulation, packaging, lysis and lysogeny. A putative Cro repressor gene and an integrase gene were found in the genome, showing that QHHSV-1 may utilize a lambda-like repression system under unfavorable conditions. QHHSV-1 is the first report of the whole genome sequence of the virulent Halomonas phage belonging to the family Siphoviridae.

Similar content being viewed by others
References
Porter K, Russ BE, Dyall-Smith ML (2007) Virus-host interactions in salt lakes. Curr Opin Microbiol 10:418–424
Adriaenssens EM, Zyl LJV, Cowan DA et al (2016) Metaviromics of Namib Desert Salt Pans: a novel lineage of Haloarchaeal Salterproviruses and a rich source of ssDNA viruses. Viruses 8:14
Oren A (2002) Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J Ind Microbiol Biotechnol 28:56–63
Atanasova NS, Bamford DH, Oksanen HM (2016) Virus–host interplay in high salt environments. Environ Microbiol Rep 8:431–444
Pedrós-Alió C, Calderón-Paz JI, Maclean MH et al (2000) The microbial food web along salinity gradients. FEMS Microbiol Ecol 32:143–155
Boujelben I, Yarza P, Almansa C et al (2012) Virioplankton community structure in Tunisian solar salterns. Appl Environ Microbiol 78:7429–7437
Atanasova NS, Oksanen HM, Bamford DH (2015) Haloviruses of archaea, bacteria, and eukaryotes. Curr Opin Microbiol 25:40–48
Fu CQ, Bai M, Wang YX et al (2017) Advances in bacteriophages isolated from hypersaline environments. Microbiol China 44(4):920−928
Vreeland RH, Litchfield CD, Martin EL et al (1980) Halomonas elongata, a new genus and species of extremely salt-tolerant bacteria. Int J Syst Bacteriol 30:485–495
Lim J-M, Yoon JH, Lee J-C et al (2004) Halomonas koreensis sp. nov., a novel moderately halophilic bacterium isolated from a solar saltern in Korea. Int J Syst Evol Microbiol 54:2037–2042
Mobberley JM, Authement RN, Segall AM et al (2008) The temperate marine phage phihap-1 of Halomonas aquamarina possesses a linear plasmid-like prophage genome. J Virol 82:6618–6630
Wang YX, Xiao W, Dong MH et al (2014) Halomonas qiaohouensis, sp. nov. isolated from salt mine soil in southwest China. Antonie van Leeuwenhoek 106:253–260
Fu CQ, Zhao Q, Li ZY et al (2016) A novel Halomonas ventosae-specific virulent halovirus isolated from the Qiaohou salt mine in Yunnan, southwest China. Extremophiles 20:101–110
Chevreux B, Pfisterer T, Drescher B et al (2004) Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res 14:1147–1159
Besemer J, Lomsadze A, Borodovsky M (2001) GenemarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618
Stephen FA, Thomas LM, Alejandro AS et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Marchler-Bauer A, Derbyshire MK, Gonzales NR et al (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43(Database issue):D222–D226
Bjellqvist B, Hughes GJ, Pasquali C et al (1993) The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14:1023–1031
Laslett D, Canback B (2004) ARAGORN, a program for the detection of transfer RNA and transfer-messenger RNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16
Becker B, de la Fuente N, Gassel M et al (1997) Head morphogenesis genes of the Bacillus subtilis bacteriophage SPP1. J MolBiol 268:822–839
Duda RL, Martincic K, Hendrix RW (1995) Genetic basis of bacteriophage HK97 prohead assembly. J Mol Biol 247:636–647
Abuladze NK, Gingery M, Tsai J et al (1994) Tail length determination in bacteriophage T4. Virology 199:301–310
Pedersen M, Ostergaard S, Bresciani J et al (2000) Mutational analysis of two structural genes of the temperate lactococcal bacteriophage TP901-1 involved in tail length determination and baseplate assembly. Virology 276:315–328
Cardarelli L, Lam R, Tuite A et al (2010) The crystal structure of bacteriophage HK97 gp6: defining a large family of head–tail connector proteins. J Mol Biol 395:754–768
Ackermann HW, DuBow MS (1987) Viruses of prokaryotes. CRC Press, Boca Raton
Ryder L, Sharples GJ, Lloyd RG (1996) Recombination-dependent growth in exonuclease-depleted recBCsbcBC strains of Escherichia coli K-12. Genetics 143:1101–1114
Mehr IJ, Long CD, Serkin CD et al (2000) A homologue of the recombination-dependent growth gene, Rdgc, is involved in Gonococcalpilin antigenic variation. Genetics 154:523–532
Sharples GJ, Chan SN, Mahdi AA et al (1995) Processing of intermediates in recombination and DNA repair: identification of a new endonuclease that specifically cleaves Holliday junctions. EMBO J 13:6133–6142
Mahdi AA, Sharples GJ, Mandal TN et al (1996) Holliday junction resolvases encoded by homologous rusA genes in Escherichia coli K-12 and phage 82. J Mol Biol 257:561–573
Waite-Rees PA, Keating CJ, Moran LS et al (1991) Characterization and expression of the Escherichia coli Mrr restriction system. J Bacteriol 173:5207–5219
King G, Murray NE (1994) Restriction enzymes in cells, not eppendorfs. Trends Microbiol 2:465–469
Bazinet C, King J (1985) The DNA translocating vertex of dsDNA bacteriophage. Annu Rev Microbiol 39:109–129
Rao VB (2008) The bacteriophage DNA packaging motor. Annu Rev Genet 42:647–681
Baxevanis AD (2005) In: Baxevanis AD, Ouellette BFF (eds) Bioinformatics. Wiley, Hoboken, pp 295–324
Claverie J-M, Notredame C (2007) Bioinformatics for dummies. Wiley Publishing Inc, Indianapolis
Krupovic M, Forterre P, Bamford DH (2010) Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J Mol Biol 397:144–160
Pietilä MK, Laurinmäki P, Russell DA et al (2013) Insights into head-tailed viruses infecting extremely halophilic archaea. J Virol 87:3248–3260
Garcia DL, Dillard JP (2006) AmiC functions as an N-acetylmuramyl-l-alanine amidase necessary for cell separation and can promote autolysis in Neisseria gonorrhoeae. J Bacteriol 188:7211–7221
Fattah KR, Mizutani S, Fattah FJ et al (2000) A comparative study of the immunity region of lambdoid phages including Shiga-toxin-converting phages: molecular basis for cross immunity. Genes Genet Syst 75:223–232
Ptashne M (2004) A genetic switch: phage lambda revisited. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Lehours P, Vale FF, Bjursell MK et al (2011) Genome sequencing reveals a phage in Helicobacter pylori. Mbio 2:1867–1877
Mei Y, He C, Huang Y et al (2015) Salinity regulation of the interaction of halovirus SNJ1 with its host and alteration of the halovirus replication strategy to adapt to the variable ecosystem. PLoS One 10:537–542
Aalto AP, Bitto D, Ravantti JJ et al (2012) Snapshot of virus evolution in hypersaline environments from the characterization of a membrane-containing salisaeta icosahedral phage 1. Proc Natl Acad Sci USA 109:7079–7084
Casjens SR, Gilcrease EB, Huang WM et al (2004) The pKO2 linear plasmid prophage of Klebsiella oxytoca. J Bacteriol 186:1818–1832
Ravin V, Ravin N, Casjens S et al (2000) Genomic sequence and analysis of the atypical temperate bacteriophage N15. J Mol Biol 299:53–73
Acknowledgements
This work was funded by grants from the National Natural Science Foundation of China (31660042, 31200138, 31660001 and 31660089), the National Infrastructure of Natural Resources for Science and Technology Program of China (NIMR-2016-8) and the Yunnan Provincial Sciences and Technology Department (2014FB104, 2009CD012) and the Yunnan University (2009C14Q).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fu, C., Zhao, Q., Li, Z. et al. Complete genome sequence of Halomonas ventosae virulent halovirus QHHSV-1. Arch Virol 162, 3215–3219 (2017). https://doi.org/10.1007/s00705-017-3415-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3415-0