Abstract
Potato virus M (PVM, genus Carlavirus, family Betaflexviridae) is considered to be one of most economically important pathogens of pepino in China. However, the details and the mechanisms underlying PVM evolution are unknown. In this study, we determined and analyzed 40 TGB 1 gene sequences, 67 TGB 2 and TGB 3 gene sequences, and 88 CP and NABP gene sequences from viruses isolated from 19 samples of pepino (Solanum muricatum) and one sample of tomato (S. lycopersicum) collected from different areas of China. Recombination analysis identified only one clear recombinant in the TGB2-TGB3-CP region, but no recombinants were detected for each of the five individual genes. Phylogenetic analysis showed that all PVM isolates could be divided into at least two lineages in trees derived from the TGB 2, CP, and NABP gene sequences, and the lineages seemed to reflect geographical origin. The five PVM genes in this study were found to be under strong negative selection pressure. The PVM isolates examined showed frequent gene flow between the Chinese and European populations, and also within the Chinese population. Clear star phylogenies and the neutral equilibrium model test showed that pepino isolates of PVM appear to be experiencing a new expansion after a recent introduction into China, and these isolates display low levels of genetic diversity. To our knowledge, this study is the first report describing genetic structure, recombination, and gene flow in PVM populations, and it provides strong evolutionary evidence for the virus populations from different geographic regions of China.





Similar content being viewed by others
References
Alabi OJ, Martin RR, Naidu RA (2010) Sequence diversity, population genetics and potential recombination events in grapevine rupestris stem pitting-associated virus in Pacific North-West vineyards. J Gen Virol 91:265–276
Briddon RW, Stanley J (2006) Subviral agents associated with plant single-stranded DNA viruses. Virology 344:198–210
Chare ER, Holmes EC (2004) Selection pressures in the capsid genes of plant RNA viruses reflect mode of transmission. J Gen Virol 85:3149–3157
Dolby CA, Jones RAC (1988) The relationship between the Andean strain of potato virus S and pepino latent virus. Ann Appl Biol 112:233–234
Domingo E, Biebricher C, Eigen M, Holland JJ (2001) Quasispecies and RNA virus evolution: principles and consequences. Landes Bioscience, Austin
Domingo E, Wain-Hobson S (2009) The 30th anniversary of quasispecies. Meeting on ‘Quasispecies: past, present and future’. EMBO Rep 10:444–448
Eigen M, McCaskill J, Schuster P (1988) Molecular quasispecies. J Phys Chem 92:6881–6891
Flatken S, Ungewickell V, Menzel W, Maiss E (2008) Construction of an infectious full-length cDNA clone of potato virus M. Arch Virol 153:1385–1389
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
García-Arenal F, Fraile A, Malpica JM (2001) Variability and genetic structure of plant virus populations. Ann Rev Phytopathol 39:157–186
Ge BB, He Z, Jiang DM, Zhang ZX, Liu GJ, Wang HQ (2012) Characterization and complete nucleotide sequence of potato virus M isolated from tomato in China. Acta Virol 56:261–263
Ge BB, Li Q, Liu GJ, Lu MG, Wang HQ, Li SF (2013) Simultaneous detection and identification of four virus infecting pepino by multiplex RT-PCR. Arch Viol 158:1181–1187
Gibbs MJ, Armstrong JS, Gibbs AJ (2000) Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16:573–582
Glasa M, Palkovics L, Kominek P, Labonne G, Pittnerova S, Kudela O, Candresse T, Subr Z (2004) Geographically and temporally distant natural recombinant isolates of Plum pox virus (PPV) are genetically very similar and form a unique PPV subgroup. J Gen Virol 85:2671–2681
Gramstat A, Courtpzanis A, Rhode W (1990) The 12 kDa protein of potato virus M displays properties of a nucleic acid binding protein. FEBS Lett 276:34–38
Hall TA (1999) BIOEDIT: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nuc Acids Symp Ser 41:95–98
Holmes EC, Ghedin E, Miller N, Taylor J, Bao Y, Georage KS, Grenfell BT, Salzberg SL, Fraser CM, Lipman DJ, Taubenberger JK (2005) Whole genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol 3:e300
Hudson RR (2000) A new statistic for detecting genetic differentiation. Genetics 155:2011–2014
Jones RAC, Koening R, Lesemann DE (1980) Pepino mosaic virus, a new potexvirus from pepino (Solanum muricatum). Ann Appl Biol 94:61–68
Ju HJ, Samuels TD, Wang YS, Blancaflor E, Payton M, Mitra R, Krishnamurthy K, Nelson RS (2005) The potato virus X TGBp2 movement protein associates with endoplasmic reticulum-derived vesicles during virus infection. Plant Physiol 138:1877–1895
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Koonin EV, Dolja VV (1993) Evolution and taxonomy of positive-strand RNA viruses: Implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol 28:375–430
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) CLUSTAL W and CLUSTAL X version 2.0. Bioinformatics 23:2947–2948
Lauring AS, Andino R (2010) Quasispecies theory and the behavior of RNA viruses. PLos Pathol 6:e1001005
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Martin D, Rybicki E (2000) RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563
Martin D, Williamson C, Posada D (2005) RDP2: recombination detection and analysis from sequence alignment. Bioinformatics 21:260–262
Maynard SJ (1992) Analyzing the mosaic structure of genes. J Mol Evol 34:126–129
Moreno IM, Malpica JM, Díaz-Pendón JA, Moriones E, Fraile A, García-Arenal F (2004) Variability and genetic structure of the population of watermelon mosaic virus infecting melon in Spain. Virology 318:451–460
Morozov SY, Miroshnichenko NA, Solovyev AG, Zelenina DA, Fedorkin ON, Lukasheva LI, Grachev SA, Chernov BK (1991) Evidence for two nonoverlapping functional domains in the potato virus X 25 K movement protein. Virology 183:782–785
Nguyen HD, Tran HT, Ohshima K (2013) Genetic variation of the Turnip mosaic virus population of Vietnam: a case study of founder, regional and local influences. Virus Res 171:138–149
Ogawa T, Tomitaka Y, Nakagawa A, Ohshima K (2008) Genetic structure of a population of Potato virus Y inducing potato tuber necrotic ringspot disease in Japan; comparison with North American and European populations. Virus Res 131:199–212
Ohshima K, Yamaguchi Y, Hirota R, Hamamoto T, Tomimura K, Tan Z, Sano T, Azuhata F, Walsh JA, Fletcher J, Chen J, Gera A, Gibbs A (2002) Molecular evolution of Turnip mosaic virus: evidence of host adaptation, genetic recombination and geographical spread. J Gen Virol 83:1511–1521
Ojosnegrosa S, Peralesb C, Masd A, Domingo E (2011) Quasispecies as a matter of fact: viruses and beyond. Virus Res 162:203–215
Page RDM (1996) Treeview: an application to display phylogenetic trees on personal computer. CABIOS 12:357–358
Pamilo P, Bianchi N (1993) Evolution of the Zfx and Zfy genes: Rates and Interdependence between the genes. Mol Biol Evol 10:271–281
Pérez-Benlloch L, Prohens J, Soler S, Nuez F (2001) Yield and fruit quality losses caused by ToMV in pepino (Solanum muricatum L.) and search for sources of resistance. Euphytica 120:24–256
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256
Posada D, Crandall KA (2011) Evaluation of methods for detecting recombination from DNA sequences: computer simulations. PNAS 98:13757–13762
Rabaa MA, Ty Hang VT, Wills B, Farrar J, Simmons CP, Holmes EC (2010) Phylogeography of recently emerged DENV-2 in southern Viet Nam. PLoS Negl Trop Dis 4:e766
Rozanov MN, Koonin EV, Gorbalenya AE (1992) Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J Gen Virol 73:2129–2134
Salminen MO, Carr JK, Burke DS, McCutchan FE (1995) Identification of breakpoints in intergenotypic recombinants of HIV type 1 by Bootscanning. AIDS Res Hum Retrov 11:1423–1425
Sawyer SA (1999) Geneconv: a computer package for the statistical detection of gene conversion. Distributed by the Author. Department of Mathematics. Washington University in St Louis. Available at http://www.math.wustl.edu/sawyer
Senshu H, Yamaji Y, Minato N, Shiraishi T, Maejima K, Hashimoto M, Miura C, Neriya Y, Namba S (2011) A dual strategy for the suppression of host antiviral silencing: two distinct suppressors for viral replication and viral movement encoded by potato virus M. J Virol 85:10269–10278
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tomimura K, Spak J, Katis N, Jenner CE, Walsh JA, Gibbs AJ, Ohshima K (2004) Comparisons of the genetic structure of populations of Turnip mosaic virus in West and East Eurasia. Virology 330:408–423
Tomitaka Y, Ohshima K (2006) A phylogeographical study of the Turnip mosaic virus population in East Asia reveals an ‘emergent’ lineage in Japan. Mol Ecol 15:4437–4457
Traore O, Sorho F, Pinel A, Abubakar Z, Banwo O, Maley J, Hebrard E, Winter S, Sere Y, Konate G, Fargette D (2005) Processes of diversification and dispersion of Rice yellow mottle virus inferred from large-scale and high-resolution phylogeographical studies. Mol Ecol 14:2097–2110
Tsompana M, Abad J, Purugganan M, Moyer JW (2005) The molecular population genetics of the Tomato spotted wilt virus (TSWV) genome. Mol Ecol 14:53–66
Wei TY, Yang JG, Liao FL, Gao FL, Lu LM, Zhang XT, Li F, Wu ZJ, Lin QY, Xie LH (2009) Genetic diversity and population structure of rice stripe virus in China. J Gen Virol 90:1025–1034
Weiller GF (1998) Phylogenetic profiles: a graphical method for detecting genetic recombinations in homologous sequences. Mol Biol Evol 15:326–335
Xu HM, Aubin JD, Nie JB (2010) Genomic variability in Potato virus M and the development of RT-PCR and RFLP procedures for the detection of this virus in seed potatoes. Virol J 7:25
Zavriev SK, Kanyuka KV, Levay KE (1991) The genome organization of potato virus M RNA. J Gen Virol 72:9–14
Zhang CL, Guo R, Wang J, Zhang GM, Li XD, Liu HT (2011) Molecular variability of Tobacco vein banding mosaic virus populations. Virus Res 158:188–198
Acknowledgments
This work was supported by grants from the Earmarked Fund for China Agriculture Research System (CARS-31), the State Scholarship Fund (No. 201203250024), the Special Fund for Agro-Scientific Research in the Public Interest (Nos. 201203076 and 200903004), the National Basic Research and Development Program of China (973 Program) (No. 2009CB119200) and the National Natural Science Foundation of China (Nos. 31401796, 31171819 and 31000842).
Author information
Authors and Affiliations
Corresponding authors
Additional information
B. Ge and Z. He contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2014_2180_MOESM1_ESM.doc
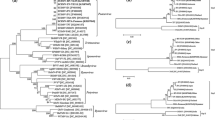
Figure S1. Maximum-likelihood trees calculated from the TGB1 (A) and TGB3 (B) gene sequences of potato virus M obtained in this study. Numbers at each node indicate the percentage of supporting puzzling steps (or bootstrap samples) in the maximum-likelihood and neighbor-joining trees. The horizontal branch length is drawn to scale, with the bar indicating 0.1 nt replacements per site. Isolate names shown in red indicate the PVM isolate sequences determined in this study. (DOC 8388 kb)
Rights and permissions
About this article
Cite this article
Ge, B., He, Z., Zhang, Z. et al. Genetic variation in potato virus M isolates infecting pepino (Solanum muricatum) in China. Arch Virol 159, 3197–3210 (2014). https://doi.org/10.1007/s00705-014-2180-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-014-2180-6