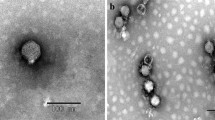
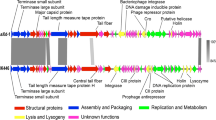
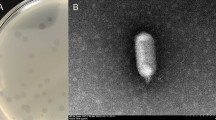
Abstract
Bacteriophage chi is a well-known phage that infects pathogens such as E. coli, Salmonella, and Serratia via bacterial flagella. To further understand its host-phage interaction and infection mechanism via host flagella, the genome was completely sequenced and analyzed. The phage genome contains 59,407-bp-length DNA with a GC content of 56.51 %, containing 75 open reading frames (ORFs) with no tRNA genes. Its annotation and functional analysis revealed that chi is evolutionarily very closely related to Enterobacter phage Enc34 and Providencia phage Redjac. However, most of the annotated genes encode hypothetical proteins, indicating that further genomic study of phage chi is required to elucidate the bacterial-flagellum-targeting infection mechanism of phage chi.


Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Barbara G, Stanghellini V, Berti-Ceroni C, De Giorgio R, Salvioli B, Corradi F, Cremon C, Corinaldesi R (2000) Role of antibiotic therapy on long-term germ excretion in faeces and digestive symptoms after Salmonella infection. Aliment Pharmacol Ther 14:1127–1131
Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618
Bruttin A, Brussow H (2005) Human volunteers receiving Escherichia coli phage T4 orally: a safety test of phage therapy. Antimicrob Agents Chemother 49:2874–2878
Cairns BJ, Payne RJH (2008) Bacteriophage therapy and the mutant selection window. Antimicrob Agents Chemother 52:4344–4350
Calendar R (2006) The bacteriophages, 2nd edn. Oxford University Press, Oxford
Campbell A (2003) The future of bacteriophage biology. Nat Rev Genet 4:471–477
Carver T, Berriman M, Tivey A, Patel C, Bohme U, Barrell BG, Parkhill J, Rajandream MA (2008) Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676
Casjens SR, Gilcrease EB (2009) Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol Biol 502:91–111
CDC (2007) Bacterial foodborne and diarrheal disease national case surveillance: annual report, 2005. Centers for Disease Control and Prevention, Atlanta
CDC (2008) Salmonella surveillance: annual summary, 2006. Centers for Disease Control and Prevention, Atlanta
Coffey B, Mills S, Coffey A, McAuliffe O, Ross RP (2010) Phage and their lysins as biocontrol agents for food safety applications. Ann Rev Food Sci Technol 1:449–468
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679
Karmali MA, Steele BT, Petric M, Lim C (1983) Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet 1:619–620
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lindberg AA (1973) Bacteriophage receptors. Ann Rev Microbiol 27:205–241
McNair K, Bailey BA, Edwards RA (2012) PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 28:614–618
Mead PS, Griffin PM (1998) Escherichia coli O157:H7. Lancet 352:1207–1212
Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV (1999) Food-related illness and death in the United States. Emerg Infect Dis 5:607–625
Meynell EW (1961) A phage, øχ, which attacks motile bacteria. J Gen Microbiol 25:253–290
Payne RJH, Jansen VAA (2000) Phage therapy: The peculiar kinetics of self-replicating pharmaceuticals. Clin Pharmacol Ther 68:225–230
Sambrook J, Russell D (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, NY
Samuel ADT, Pitta TP, Ryu WS, Danese PN, Leung ECW, Berg HC (1999) Flagellar determinants of bacterial sensitivity to χ-phage. P Natl A Sci USA 96:9863–9866
Sertic V, Boulgakov N-A (1936) Bactnriophages specifique pour des varites bacteriennes flagellees. C R Soc Biol Paris 113:105–113
Wilcox SA, Toder R, Foster JW (1996) Rapid isolation of recombinant lambda phage DNA for use in fluorescence in situ hybridization. Chromosome Res 4:397–398
Yamaguchi S, Fujita H, Sugata K, Taira T, Iino T (1984) Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J Gen Microbiol 130:255–265
Zdobnov EM, Apweiler R (2001) InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848
Acknowledgments
This research was supported by WCU (World Class University) program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (R32-2008-000-10183-0) and the R&D Convergence Center Support Program of the Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lee, JH., Shin, H., Choi, Y. et al. Complete genome sequence analysis of bacterial-flagellum-targeting bacteriophage chi. Arch Virol 158, 2179–2183 (2013). https://doi.org/10.1007/s00705-013-1700-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-013-1700-0