Abstract
Bovine torovirus (BToV) is recognized as an enteric pathogen of calves, but its etiological role in diarrhea and epidemiological characterization in adult cows remain unclear. In 2007-2008, three outbreaks of epidemic diarrhea occurred in adult cows at three dairy farms in Niigata Prefecture, Japan. BToV was the only enteric pathogen detected in these outbreaks, as determined by electron microscopy, reverse transcription-PCR, bacteria and parasite tests of fecal samples, and antibody tests with paired sera. The epidemiological features of the three outbreaks were similar to those of bovine coronavirus infection, except for the absence of bloody diarrhea, with diarrhea spreading among most adult cows, but not in calves, within several days and diarrhea lasting for 3-5 days with anorexia. Decreased milk production and mild respiratory symptoms were also observed in two of the outbreaks. Nucleotide sequence analysis of the BToV nucleocapsid, spike, and hemagglutinin-esterase (HE) genes revealed a close relatedness among the detected BToV strains from each outbreak and those of Japanese BToV strain Aichi/2004. Furthermore, we isolated a BToV strain, designated Niigata (TC), from a fecal sample using a human rectal tumor cell line. Sequence analysis of this isolate and Aichi/2004 indicated that both strains have truncated HE genes with deletions in the 3′ region that occurred through cell culture-adaptation. The short projections that are believed to be formed by the HE protein on virus particles were not observed in these cultured strains by electron microscopy. Taken together, these results suggest that BToV causes epidemic diarrhea in adult cows and should be included in the differential diagnosis of diarrhea in adult cows. In addition, our findings indicate that the HE protein of BToV may not be necessary for viral replication.
Similar content being viewed by others
Introduction
Epidemic diarrhea in adult cattle is frequently observed worldwide, particularly among dairy cattle, and results in large economic losses from marked reductions in milk production. A major causative agent of adult cattle diarrhea is bovine coronavirus (BCV) [19, 24], which leads to the disease winter dysentery (WD). However, other viruses, including bovine torovirus (BToV), are also suspected to be etiologic agents of diarrhea [14, 15, 21], although the epidemiological situation of infections caused by these viruses in adult cattle remains unclear.
BToV, a member of the genus Torovirus within the family Coronaviridae, is a spherical, oval, elongated, or kidney-shaped enveloped virus and displays a double fringe of peplomers on its surface [3]. The virus has a single-stranded RNA genome of positive polarity encoding RNA polymerase and four structural proteins: the spike (S), membrane (M), hemagglutinin-esterase (HE), and nucleocapsid (N) proteins [5]. Since the first isolation of BToV in 1982 from a case of neonatal calf diarrhea in the United States [34], several reports have confirmed the association of BToV with calf diarrhea in experimentally infected gnotobiotic calves and under field conditions [6, 8, 9, 13, 18]. Serological surveys have also revealed a high prevalence of antibodies to BToV in cattle populations, ranging between 55% and 95% [2, 14, 16, 33, 35]. Recently, Kuwabara et al. [16] succeeded in isolating BToV using a human rectal tumor cell line (HRT-18), which is expected to facilitate the diagnosis of BToV infection by virus neutralization tests and virus isolation.
BToV has been detected in fecal samples from adult cattle with diarrhea [10, 11, 21]. In addition, a few studies have reported that adult cows with WD-like diarrhea exhibited various degrees of seroconversion to BToV [14, 15]. These results suggest that BToV may be a causative agent of epidemic diarrhea of adult cattle. However, the epidemiological data for BToV infection in adult cattle under field conditions are limited, and the etiologic role of BToV in epidemic diarrhea of adult cattle remains unclear.
In the present study, we report the epidemiological features of three outbreaks of epidemic diarrhea in adult cows between May 2007 and February 2008 at three dairy farms in Niigata Prefecture, Japan, in which BToV was detected as the only pathogen. Further, we describe the isolation of a cytopathogenic BToV strain from diarrheic feces using HRT-18 cells, as well as the molecular characterization of the detected BToVs.
Materials and methods
Clinical specimens
Three epidemic outbreaks of adult cattle diarrhea occurred between May 2007 and February 2008 in Niigata Prefecture, Japan. A total of 16 fecal samples were collected from 16 diarrheic adult cows aged 24 to 71 months: 5 samples (Niigata1-1 to Niigata1-5) from outbreak 1 (Farm 1), 7 samples (Niigata2-1 to Niigata2-7) from outbreak 2 (Farm 2), and 4 samples (Niigata3-1 to Niigata3-4) from outbreak 3 (Farm 3). Nasal swabs were also collected at the time of fecal sampling from the 7 cows involved in outbreak 2. Paired serum samples were collected in the acute phase of diarrhea and 14 to 19 days later. Fecal samples were diluted 1:10 in 0.01 M phosphate-buffered saline (PBS; pH 7.4), and nasal swabs were suspended in 2 ml PBS. The resulting suspensions were clarified by low-speed centrifugation at 3,000×g for 10 min and used for virus isolation and reverse transcription-polymerase chain reaction (RT-PCR). Fecal samples were also tested for Salmonella species using a standard technique, while Coccidium species and Cryptosporidium species were detected by a sucrose floatation method. In addition, fecal samples were examined for rotavirus double-stranded RNA by polyacrylamide gel electrophoresis [17].
Virus-neutralization and hemagglutination inhibition tests
Virus neutralizing antibody titers against BToV in paired sera from affected cows were measured using HRT-18 cells and the BToV strain Aichi/2004 [16]. Virus neutralization (VN) tests were also conducted for bovine viral diarrhea (BVDV) type 1, bovine adenovirus type 3 (BAdV-3), bovine herpesvirus 1 (BHV-1), and bovine respiratory syncytial virus (BRSV) as described previously [36]. Antibody titers against BCV, adenovirus type 7 (BAdV-7), and bovine parainfluenza virus type 3 (BPIV-3) were determined by hemagglutination inhibition (HI) tests [20]. Seroconversion was defined as a fourfold or greater increase in paired serum antibody titers to the examined virus.
Electron microscopy
Fecal suspensions and the supernatants of infected cell cultures were partially purified by ultracentrifugation through a 30% (wt/wt) sucrose cushion, negatively stained with 2% sodium phosphotungstic acid (pH 7.0), and examined using an electron microscope (JEM-100S; JEOL, Ltd., Tokyo, Japan).
Virus isolation
Two lines of human rectal tumor (HRT-18) and Madin-Darby bovine kidney (MDBK) cells were used for virus isolation. One line of HRT-18 cells (designated HRT-18 Aichi cells), which had been maintained in Aichi Prefecture, Japan, was used for the first isolation of BToV in cell culture [16], and the second HRT-18 cell line (designated HRT-18 Niigata cells) had been maintained in our laboratory. Confluent monolayers of these cells in 24-well plates were washed with Eagle’s MEM (EMEM) and then inoculated with 0.1 ml of the fecal suspensions diluted from 1:10 to 1:1000 in EMEM with and without 10 μg/ml trypsin (Type I; Sigma-Aldrich, St. Louis, MO, USA). After adsorption for 60 min at 37°C, the cells were washed once with EMEM, 0.5 ml EMEM was added, and they were further incubated for 4 or 5 days at 37°C and examined for cytopathic effects (CPE). After incubation, the cells and culture supernatant were frozen and thawed once to harvest cell lysates, and subsequent passages were carried out in the same manner with 0.1 ml of cell lysate. After two passages, the viral isolate was cloned three times in HRT-18 cells by limiting dilution.
The isolate was identified by indirect immunofluorescence and VN tests with gnotobiotic calf antisera against BToV (kindly supplied by Dr. Linda Saif, The Ohio State University) [9] and BCV as described previously [16, 28]. For VN tests, BToV strain Aichi/2004 [16] and BCV strain Kakegawa [1] were used as reference viruses. Electron microscopy and RT-PCR were also performed for the identification of the isolate.
RT-PCR
Viral RNA was extracted from fecal suspensions, nasal swabs, and infected cell culture supernatants using TRIzol LS Regent (Invitrogen Corp., Carlsbad, CA, USA) according to the manufacturer’s instructions. RT-PCR assays were performed using a OneStep RT-PCR kit (QIAGEN, Valencia, CA, USA) and primer pairs targeting fragments of the N-, S-, and HE-specific genes of torovirus. The primers, which were identical to those described previously by Hoet et al. [10] and Smits et al. [23], were as follows: 1344 (5′-GAGAAAGAGCCAAGATGAATT-3′; positions 27761 to 27781) and 294 (5′-CTTACATGGAGACACTCAACCA-3′; positions 28403 to 28424) for N genes, primers S5 (5′-GTGTTAAGTTTGTGCAAAAAT-3′; positions 20956 to 20976) and S3 (5′-TGCATGAACTCTATATGGTGT-3′; positions 21677 to 21697) for S genes, and primers 1433 (5′-AAGTTTGAGTAGCCACTTATC-3′; positions 26448 to 26468) and 1383 (5′-TAGCATTTGGATTAAGCATAG A-3′; positions 27781 to 27801) for HE genes. The positions were based on the genome of BToV strain Breda1 (AY427798). RT-PCR was also performed for detection of the specific genes of BCV [30], group A rotavirus (GAR) [7], group B rotavirus (GBR) [4], group C rotavirus (GCR) [29], and BVDV [32].
Sequence analysis
PCR products of the N, S, and HE genes of BToV were sequenced directly by cycle sequencing with an automatic sequencer (ABI PRISM 3100; Applied Biosystems, Tokyo, Japan). Sequencing was also performed for the HE gene of strain Aichi/2004 [16], which was passaged nine times in HRT-18 Aichi cells, and for the N, S, and HE genes from the large-intestinal content of the original specimen from which Aichi/2004 was isolated, designated as Aichi/2004(LIC). Sequence data were analyzed by the Clustal W method [26] using the Megalign 7.2 program of Lasergene software (DNASTAR, Madison, WI, USA). Phylogenetic trees were constructed using the MEGA 4 program [25].
Nucleotide sequence accession numbers
The newly determined sequences have been deposited in the DDBJ nucleotide sequence database and assigned the following accession numbers: Niigata1 (N gene, AB661450; S gene, AB661453; HE gene, AB661456), Niigata2 (N gene, AB661451; S gene, AB661454; HE gene, AB661457), Niigata3 (N gene, AB661452; S gene, AB661455; HE gene, AB661458), Niigata3(TC) (HE gene, AB661459), Aichi/2004(LIC) (HE gene, AB661461), and Aichi/2004 (HE gene, AB661460).
Results
Epidemiological situation of diarrhea outbreaks
Outbreak 1
On May 16, 2007, diarrhea and anorexia were observed in a few adult lactating cows on a dairy farm (Farm 1), with 12 of 24 adult cows on the farm displaying similar symptoms within several days. The diarrheal feces were watery and brownish. All affected cows recovered from diarrhea within 4-5 days, but milk production had decreased by up to 7% on the second day following the first finding of diarrhea and continued for one week.
Outbreak 2
On November 24, 2007, a few adult lactating cows on a dairy farm (Farm 2) showed pasty or watery diarrhea that was accompanied by nasal discharge. Within 4 days, 15 of 47 adult cows showed diarrhea and most of them had nasal discharge. Milk production decreased by approximately 20% on the fourth day after the onset of diarrhea. All affected cows recovered from diarrhea within 4-5 days. The farm was located approximately 80 km from Farm 1. No epidemiological relationship was observed between Farms 1 and 2.
Outbreak 3
On February 15, 2008, symptoms of pasty or watery diarrhea, anorexia, and mild cough were observed in a few adult lactating cows on a dairy farm (Farm 3), located 2 km away from Farm 1. Within one week, all 22 adult cows on the farm showed similar clinical signs. All cows recovered from diarrhea after 3-4 days, and no clinical signs were observed on the tenth day after the first finding of diarrhea. Milk production did not change during the outbreak.
Several common epidemiological features were observed among the three outbreaks. Affected cows did not show bloody diarrhea or fever, and each cow recovered within 3-5 days after the onset of diarrhea without clinical treatment. Notably, no clinical signs, including diarrhea, were observed in calves during these outbreaks of adult cow diarrhea.
Examination of fecal samples
All 16 fecal samples were RT-PCR negative for BCV, BVDV, GAR, GBR, and GCR, and were also negative for species of Salmonella, Coccidium, and Cryptosporidium. In addition, none of the samples were positive for rotavirus RNAs by polyacrylamide gel electrophoresis. In contrast, all fecal samples except for Niigta2-2 were positive for N, S, and HE genes of BToV by RT-PCR. One nasal swab sample from outbreak 2 was also positive for the BToV N gene by RT-PCR, while we could not detect the S and HE genes of BToV in any nasal samples.
Virus antibody tests
Examination of paired sera from 14 of 16 affected cows showed seroconversion (≥4-fold increase) to BToV, and serum samples from the remaining two cows had antibody titers of ≥512 at the acute phase (Table 1). No significant change in BCV antibody titers was observed in any of the outbreaks (Table 1). Additionally, antibody titers of the other viruses examined (BVDV type 1, BAdV-3, BAdV-7, BHV-1, BRSV, and BPIV-3) did not markedly change in any of the paired sera.
Virus isolation in HRT-18 Aichi cells
After the second passage, CPE appeared in HRT-18 Aichi cells inoculated with one fecal sample, Niigata3-4, diluted 1:100 with EMEM containing trypsin. CPE, which was characterized by the enlargement of cells, was observed 2 to 3 days after inoculation, with the cells eventually detaching from the plastic surface. The isolate was cloned by limiting dilution three times, and the infective titers reached 105.5 TCID50/ml by passage 7. Indirect immunofluorescence microscopy of HRT-18 Aichi cells that had been inoculated with the isolate and reacted with gnotobiotic calf antiserum against BToV revealed cytoplasmic fluorescence, indicating the presence of BToV antigens [16]. In contrast, no fluorescent cells were observed when the infected cells were reacted with anti-BCV antiserum.
In VN tests, antiserum to BToV neutralized strain Aichi/2004 and the isolate at titers of 25,600 and 12,800, respectively. In contrast, antiserum to BCV failed to neutralize either strain (titer, <50). In addition, the isolate was positive in RT-PCR targeting the N, S, and HE genes of BToV. Electron microscopy and nucleotide sequencing of the RT-PCR products were also conducted for identification of the isolate. From these results, we concluded that the isolate from fecal sample Niigata3-4 was BToV and designated the new isolate as strain Niigata3(TC). In contrast, no cytopathogenic viruses were isolated from any samples using HRT-18 Niigata or MDBK cells.
Electron microscopy
Coronavirus-like particles were observed by electron microscopy in 5 of 16 fecal samples (Niigata1-2, -3, -4; Niigata2-3, and Niigata3-2) from the three outbreaks. The particles were spherical, oval, elongated, or kidney-shaped, approximately 100-140 nm in diameter, and surrounded by club-shaped projections of 15-20 nm in length. The particles also had short projections of approximately 5 nm in length (Fig. 1a, b). No other virus-like particles were observed in any of the fecal samples.
Electron micrographs of virus particles in fecal samples from adult cows and strain Niigata3(TC) isolated using HRT-18 Aichi cells. a and b are from the fecal samples Niigata3-2 and Niigata1-4, respectively. The particles possess long (15-20 nm) and short (approximately 5 nm) projections. c and d are from the Niigata3(TC) isolate. The particles possess long, but not short, projections. Bar, 100 nm
In the culture supernatants of infected HTR-18 Aichi cells, coronavirus-like particles with long club-shaped projections similar to those detected in the fecal specimens were observed. However, short projections were not seen on these particles (Fig. 1c, d).
Analysis of BToV genes
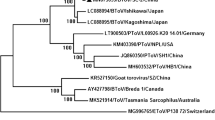
The nucleotide sequences of the N (485 nucleotides [nt]), S (673 nt), and HE genes (1260 nt) of BToV from nine fecal samples obtained from the three farms and BToV isolate Niigata3(TC) were compared with those of published toroviruses. The nine selected fecal samples were as follows: Niigata1-1, -2, and -3 from outbreak 1, Niigata2-3 and -4 from outbreak 2, and Niigata3-1, -2, -3, and -4 from outbreak 3. In addition, the nucleotide sequences of the viral genes of Aichi/2004 and Aichi/2004(LIC) were compared (Fig. 2).
Phylogenetic trees for the N (a), S (b), and HE genes (c) of toroviruses constructed using the neighbor-joining method and drawn with the MEGA 4 program. Bootstrap values greater than 700 in 1000 pseudoreplicates are shown as percentages. The accession numbers of the nucleotide sequences used for tree construction are as follows: for Aichi/2004, AB285125 (N gene), AB285127 (S gene); for Breda 1, AY427798 (N, S, and HE genes); for Breda 2, AF076621 (S and HE genes); for B6, AJ575389 (N gene) and AJ575378 (HE gene); for B145, AJ575388 (N gene), AJ575373 (S gene), and AJ575379 (HE gene); for B150, AJ575387 (N gene); for B155, AJ575386 (N gene) and AJ575381 (HE gene); for B156, AJ575385 (N gene) and AJ575382 (HE gene); for B1314, AJ575384 (N gene) and AJ575383 (HE gene); for Gifu-2009TI-E, AB526865 (S gene); for Hokkaido-2008TI-E, AB526864 (S gene); for Miyagi-2006TI-E, AB526862 (S gene); for Gifu-2007TI-E, AB526863 (S gene); for Berne, D00563 (N gene) and X52506 (S gene); for BRES, FJ232070 (HE gene); for Markelo, AJ575358 (N gene), AJ575372 (S gene), and AJ575363 (HE gene); for P4, AJ575359 (N gene) and AJ575364 (HE gene); for P9, AJ575360 (N gene) and AJ575365 (HE gene); for P10, AJ575366 (HE gene); for P78, AJ575362 (N gene) and AJ575367 (HE gene). For the accession numbers of the BToVs from the present study, please refer to the text
As the nucleotide sequences of BToV genes from the same outbreak were identical, BToV detected from the feces in each respective outbreak was designated as strains Niigata1, Niigata2, and Niigata3. For the N gene, the nucleotide sequences of Niigata1 and Niigata3 were identical to that of the B145 strain detected in the Netherlands and showed 99% nucleotide identity to Niigata2. For the S gene, Niigata2 and Niigata3 showed 99% nucleotide identity, and they displayed 96%-97% nucleotide identity to Niigata1 and Aichi/2004. For the HE gene, Niigata1, Niigata2, and Niigata3 showed 98%-99% nucleotide identity to each other and 98%-99% nucleotide identity to Aichi/2004. In addition, the nucleotide sequences of both the N and S genes between Niigata3 and Niigata3(TC) and between Aichi/2004(LIC) and Aichi/2004 showed 100% identity.
Phylogenetic analysis of the N, S, and HE genes indicated that strains Niigata1, Niigata2, and Niigata3 were grouped within the same cluster as other BToV strains, including Aichi/2004 and B145 (Fig. 2). In the HE gene, the detected open reading frames of strains Niigata1, Niigata2, Niigata3, and Aichi/2004(LIC) encoded 419 amino acids, which is identical to the number reported previously for other strains of BToV. In contrast, both cell-culture-adapted Aichi/2004 and Niigata3(TC) strains of BToV had incomplete HE open reading frames that only encoded 160 and 104 amino acids, respectively (Fig. 3).
Sequences of the HE genes of BToV detected in this study. Aichi/2004 and Niigata3(TC) are cell-culture-adapted BToV strains isolated using HRT-18 Aichi cells. All other strains were detected in fecal specimens. Open boxes indicate AUG initiation codons, and gray boxes indicate termination codons of ORFs. Accession numbers of the nucleotide sequences are indicated in the legend of Fig. 2
Discussion
BToV has been suspected to be a causative agent of adult cattle diarrhea based on its detection in diarrheic feces and because various degrees of seroconversion to BToV have been observed in outbreaks of WD-like diarrhea in adult cows [10, 11, 14, 15, 31]. However, prior to the present study, no conclusive evidence for a link between BToV and epidemic diarrhea in adult cattle had been reported. Furthermore, epidemiological characterization of these diarrheal outbreaks remained unclear. Here, we examined the etiological agents and conducted detailed serological tests with samples from three outbreaks of adult cattle diarrhea in Niigata, Japan, and found that BToV was the only causative pathogen.
In the three outbreaks examined, most affected cows developed watery diarrhea, anorexia, and decreased milk production, and additionally, some cows also showed slight respiratory signs. The observed clinical signs were similar to those encountered in WD and BCV infections. Notably, none of the calves developed diarrhea during the outbreaks, a diagnostic characteristic that is also observed in WD and BCV infections in adult cows [27, 31]. Taken together, these results suggest that adult cattle diarrhea caused by BToV may have been misdiagnosed as WD or BCV infection by clinical investigations.
We isolated the cytopathogenic BToV strain Niigata3(TC) using HRT-18 cells. Of note, a clear difference in sensitivity to BToV was observed between the two HRT-18 cell lines used in this study. Namely, the Niigata3(TC) strain could be isolated and propagated in HRT-18 Aichi cells, but not in HRT-18 Niigata cells. Similarly, strain Aichi/2004 could also not be propagated in HRT-18 Niigata cells, although both HRT-18 cell lines displayed high sensitivity to BCV (data not shown). These results indicate that the use of HRT-18 Aichi cells is critical for the in vitro propagation of BToV. Although the reason for the different sensitivity between the two HRT-18 cell lines to BToV is unknown, it is possible that the properties of the HRT-18 Aichi cells may have changed during passaging to increase BToV tropism.
BToVs have been classified into two serotypes on the basis of reactivity in the hemagglutination/HI test [35]. Recently, however, BToVs have been proposed to consist of a single serotype that includes at least two subtypes [12]. In the present study, rabbit antisera to strain Aichi/2004 showed no significant differences in virus neutralizing antibody titers between the Aichi/2004 and Niigata3(TC) strains, suggesting that these two strains belong to the same serotype. Nucleotide sequence analysis indicated that the BToV strains detected in the fecal samples from three outbreaks in Niigata prefecture were closely related to each other and to strain Aichi/2004. This finding, together with the serological results, suggests that these Japanese BToV strains may have evolved from a recent common ancestor.
Our sequencing analyses revealed that both cell-culture-adapted BToV strains, Niigata3(TC) and Aichi/2004, possessed truncated HE genes with deletions in the 3′ region that occurred through cell culture-adaptation. Electron microscopy photographs indicated that virus particles in fecal samples had long and short projections, whereas short projections were not seen on those in the culture supernatants of infected HTR-18 cells. Interestingly, in many laboratory strains of mouse hepatitis virus, the HE genes are inactivated, which apparently represents an in vitro artifact resulting from adaptation to propagation in cultured cells [37]. It is noteworthy that cell-culture-adapted equine torovirus Berne virus also possesses a truncated HE gene with deletions in the 5′ region [22]. Taken together with the data presented in this study, the HE protein in BToV may be unnecessary for viral replication. Further examination of the biological properties of the HE protein in fecal- and cell-culture-adapted strains of BToV is needed to confirm this speculation.
In conclusion, we presented the epidemiological characteristics of BToV diarrhea in adult cattle based on three outbreaks in Niigata, Japan. We propose that examination for BToV should be included in the routine diagnosis of adult cattle diarrhea.
References
Akashi H, Inaba Y, Miura Y, Sato K, Tokuhisa S, Asagi M, Hayashi Y (1981) Propagation of the Kakegawa strain of bovine coronavirus in suckling mice, rats and hamsters. Arch Virol 67:367–370
Brown DWG, Beards GM, Flewett TH (1987) Detection of breda virus antigen and antibody in humans and animals by enzyme immunoassay. J Clin Microbiol 25:637–640
Cavanagh D, Horzinek MC (1993) Genus Torovirus assigned to the Coronaviridae. Arch Virol 128:395–396
Chinsangaram J, Akita GY, Osburn BI (1994) Detection of bovine group B rotaviruses in feces by polymerase chain reaction. J Vet Diagn Invest 6:302–307
Draker R, Roper RL, Petric M, Tellier R (2006) The complete sequence of the bovine torovirus genome. Virus Res 115:56–68
Duckmanton L, Carman S, Nagy É, Petric M (1998) Detection of bovine torovirus in fecal specimens of calves with diarrhea from Ontario farms. J Clin Microbiol 36:1266–1270
Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, Fang ZY (1990) Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol 28:276–282
Haschek B, Klein D, Benetka V, Herrera C, Sommerfeld-Stur I, Vilcek Š, Moestl K, Baumgartner W (2006) Detection of bovine torovirus in neonatal calf diarrhoea in lower Austria and Styria (Austria). J Vet Med B 53:160–165
Hoet AE, Cho KO, Chang KO, Loerch SC, Wittum TE, Saif LJ (2002) Enteric and nasal shedding of bovine torovirus (Breda virus) in feedlot cattle. Am J Vet Res 63:342–348
Hoet AE, Nielsen PR, Hasoksuz M, Thomas C, Wittum TE, Saif LJ (2003) Detection of bovine torovirus and other enteric pathogens in feces from diarrhea cases in cattle. J Vet Diagn Invest 15:205–212
Ito T, Okada N, Fukuyama S (2007) Epidemiological analysis of bovine torovirus in Japan. Virus Res 126:32–37
Ito T, Katayama S, Okada N, Masubuchi K, Fukuyama S, Shimizu M (2010) Genetic and antigenic characterization of newly isolated bovine toroviruses from Japanese cattle. J Clin Microbiol 48:1795–1800
Kirisawa R, Takeyama A, Koiwa M, Iwai H (2007) Detection of bovine torovirus in fecal specimens of calves with diarrhea in Japan. J Vet Med Sci 69:471–476
Koopmans M, Van Den Boom U, Wood G, Horzinek MC (1989) Seroepidemiology of breda virus in cattle using ELISA. Vet Microbiol 19:233–243
Koopmans M, Van Wuijckhuise-Sjouke L, Schukken YH, Cremers H, Horzinek MC (1991) Association of diarrhea in cattle with torovirus infections on farms. Am J Vet Res 52:1769–1773
Kuwabara M, Wada K, Maeda Y, Miyazaki A, Tsunemitsu H (2007) First isolation of cytopathogenic bovine torovirus in cell culture from a calf with diarrhea. Clin Vaccine Immunol 14:998–1004
Pereira HG, Gouvea VS, Fialho AM (1986) A comparison of simian rotavirus SA11 preparations maintained in different laboratories. Mem Inst Oswaldo Cruz 81:389–393
Pohlenz JF, Cheville NF, Woode GN, Mokresh AH (1984) Cellular lesions in intestinal mucosa of gnotobiotic calves experimentally infected with a new unclassified bovine virus (Breda virus). Vet Pathol 21:407–417
Saif LJ, Brock KV, Redman DR, Kohler EM (1991) Winter dysentery in dairy herds: electron microscopic and serological evidence for an association with coronavirus infection. Vet Rec 128:447–449
Sato K, Inaba Y, Kurogi E, Takahashi E, Satado K, Omori T, Matsumoto M (1977) Hemagglutination by calf diarrhea coronavirus. Vet Microbiol 2:83–87
Scott FMM, Holliman A, Jones GW, Gray EW, Fitton J (1996) Evidence of torovirus infection in diarrhoeic cattle. Vet Rec 138:284–285
Sinjder Ej, den Boon JA, Horzinek MC, Spaan WJ (1991) Comparison of the genome organization of toro- and coronaviruses: evidence for two nonhomologous RNA recombination events during Berne virus evolution. Virology 180:448–452
Smits SL, Lavazza A, Matiz K, Horzinek MC, Koopmans MP, de Groot RJ (2003) Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events. J Virol 77:9567–9577
Takahashi E, Inaba Y, Sato K, Ito Y, Kurogi H, Akashi H, Satoda K, Omori T (1980) Epizootic diarrhoea of adult cattle associated with a coronavirus-like agent. Vet Microbiol 5:151–154
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tråvén M, Sundberg J, Larsson B, Niskanen R (1993) Winter dysentery diagnosed by farmers in dairy herds in central Sweden: incidence, clinical signs and protective immunity. Vet Rec 133:315–318
Tsunemitsu H, Yonemichi H, Hirai T, Kudo T, Onoe S, Mori K, Shimizu M (1991) Isolation of bovine coronavirus from feces and nasal swabs of calves with diarrhea. J Vet Med Sci 53:433–437
Tsunemitsu H, Jiang B, Saif LJ (1996) Sequence comparison of the VP7 gene encoding the outer capsid glycoprotein among animal and human group C rotaviruses. Arch Virol 141:705–713
Tsunemitsu H, Smith DR, Saif LJ (1999) Experimental inoculation of adult dairy cows with bovine coronavirus and detection of coronavirus in feces by RT-PCR. Arch Virol 144:167–175
Van Kruiningen HJ, Castellano VP, Koopmans M, Harris LL (1992) A serologic investigation for coronavirus and breda virus antibody in winter dysentery of dairy cattle in the northeastern United States. J Vet Diagn Invest 4:450–452
Vilcek S, Herring AJ, Herring JA, Nettleton PF, Lowings JP, Paton DJ (1994) Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch Virol 136:309–323
Weiss M, Steck F, Kaderli R, Horzinek MC (1984) Antibodies to berne virus in horse and other animals. Vet Microbiol 9:523–531
Woode GN, Reed DE, Runnels PL, Herrig MA, Hill HT (1982) Studies with an unclassified virus isolated from diarrheic calves. Vet Microbiol 7:221–240
Woode GN, Saif LJ, Quesada M, Winand NJ, Pohlenz JF, Gourley NK (1985) Comparative studies on three isolates of Breda virus of calves. Am J Vet Res l46:1003–1010
Xue W, Ellis J, Mattick D, Smith L, Brady R, Trigo E (2010) Immunogenicity of a modified-live virus vaccine against bovine viral diarrhea virus types 1 and 2, infectious bovine rhinotracheitis virus, bovine parainfluenza-3 virus, and bovine respiratory syncytial virus when administered intranasally in young calves. Vaccine 28:3784–3792
Yokomori K, Banner LR, Lai MMC (1991) Heterogeneity of gene expression of the hemagglutinin-esterase (HE) protein of murine coronaviruses. Virology 183:647–657
Acknowledgements
We thank N. Kawanishi, T. Miyamoto, and N. Hattori for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Aita, T., Kuwabara, M., Murayama, K. et al. Characterization of epidemic diarrhea outbreaks associated with bovine torovirus in adult cows. Arch Virol 157, 423–431 (2012). https://doi.org/10.1007/s00705-011-1183-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-011-1183-9