Abstract
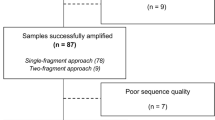
Most commercial HIV-1 genotyping assays are hampered by high cost in resource-limited settings. Moreover, their performance might be influenced over time by HIV genetic heterogeneity and evolution. An in-house genotyping protocol was developed, and its sequencing performance and reproducibility were compared to that of ViroSeq™. One hundred ninety plasma samples from HIV-1-infected subjects in Cameroon, a resource-limited setting with a high HIV genetic variability, were processed for pol gene sequencing with an in-house protocol, ViroSeq™, or both. Only non-B subtypes were found. The in-house sequencing performance was 98.7% against 92.1% with ViroSeq™. Among 36 sequence pairs obtained using both assays, the overall rate of discordant amino acid positions was negligible (0.24%). With its high sensitivity and reproducibility, as well as its affordable cost (about half of ViroSeq™: 92 € vs. 217 €), this in-house assay is a suitable alternative for HIV-1 genotyping in resource-limited and/or in high-genetic-diversity settings.

Similar content being viewed by others
References
World Health Organization (2009) Aids epidemic updates December 2009
WHO/UNAIDS (2003) The WHO and UNAIDS global initiative to provide antiretroviral therapy to 3 million people with HIV/AIDS in developing countries by the end of 2005. http://www.whoint/hiv
World Health Organization (2006) Antiretroviral therapy for HIV infection in adults and adolescents in resource-limited settings: towards universal access: recommendations for a public health approach. http://wwwwhoint/hiv/pub/guidelines/adult/en/indexhtml
Braitstein P, Brinkhof MW, Dabis F, Schechter M, Boulle A, Miotti P, Wood R, Laurent C, Sprinz E, Seyler C, Bangsberg DR, Balestre E, Sterne JA, May M, Egger M (2006) Mortality of HIV-1-infected patients in the first year of antiretroviral therapy: comparison between low-income and high-income countries. Lancet 367:817–824
Bennett DE (2006) The requirement for surveillance of HIV drug resistance within antiretroviral roll out in the developing world. Curr Opin Infect Dis 19:607–614
European Guidelines Group for HIV Resistance (2009) Clinical and laboratory guidelines for the use of HIV-1 drug resistance testing as part of treatment management: recommendations for the European setting. AIDS 15:309–320
Panel on Antiretroviral Guidelines for Adults and Adolescents Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Department of Health and Human Services December 1, 2009, pp 1–161. http://www.aidsinfonihgov/ContentFiles/AdultandAdolescentGL.pdf
Laurent C, Kouanfack C, Koulla-Shiro S, Njoume M, Nkene YM, Ciaffi L, Brulet C, Peytavin G, Vergne L, Calmy A, Mpoudi-Ngole E, Delaporte E (2007) Long-term safety, effectiveness and quality of a generic fixed-dose combination of nevirapine, stavudine and lamivudine. AIDS 21:768–771
Boyer S, Eboko F, Camara M, Abé C, Nguini ME, Koulla-Shiro S, Moatti JP (2010) Scaling up access to antiretroviral treatment for HIV infection: the impact of decentralization of healthcare delivery in Cameroon. AIDS 24(Suppl 1):S5–S15
Ndongmo CB, Pieniazek D, Holberg-Petersen M, Holm-Hansen C, Zekeng L, Jeansson SL, Kaptue L, Kalish ML (2006) HIV Genetic diversity in Cameroon: possible public health importance. AIDS Res Hum Retroviruses 22:812–816
Vallari A, Bodelle P, Ngansop C, Makamche F, Ndemb N, Mbanya D, Kaptue L, Gurtler L, McArthur C, Devare S (2010) Four new HIV-1 group N isolates from Cameroon: prevalence continues to be low. AIDS Res Hum Retroviruses 26:109–115
Plantier JC, Leoz M, Dickerson JE, de Oliveira F, Cordonnier F, Lemée V, Damond F, Robertson DL, Simon F (2009) A new human immunodeficiency virus derived from gorillas. Nat Med 15:871–872
Comité National de lutte contre le SIDA, Ministère de la Santé Publique du Cameroun July 2009 Profil des estimations pays du VIH SIDA au Cameroun
Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, Heneine W, Kantor R, Jordan MR, Schapiro JM, Vandamme AM, Sandstrom P, Boucher CA, van de Vijver D, Rhee SY, Liu TF, Pillay D, Shafer RW (2009) Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 4(3):e4724
Gallant JE (2006) Antiretroviral drug resistance and resistance testing. Top HIV Med 13:138–142
Johnson VA, Brun-Vezinet F, Clotet B, Gunthard HF, Kuritzkes DR, Pillay D, Schapiro JM, Richman DD (2009) Update of the drug resistance mutations in HIV-1. Top HIV Med 63:585–592
Tommy F, Liuand R, Shafer W (2006) Web resources for HIV type 1 genotypic resistance test interpretation. Clin Infect Dis 42:1608–1618
Mukaide M, Sugiura W, Matuda M, Usuku S, Noguchi Y, Suzuki K, Kawata K, Ito A, Sagara H, Yamada K, Kondo M, Imai M (2000) Evaluation of ViroSeq™ HIV version 2 for HIV drug resistance. Jpn J Infect Dis 53:203–205
Mracna M, Becker-Pergola G, Dileanis J, Guay LA, Cunningham S, Jackson JB, Eshleman SH (2001) Performance of the applied biosystems ViroSeq™ HIV-1 genotyping system for sequence-based analysis of non-subtype b HIV-1 from Uganda. J Clin Microbiol 39:4323–4327
Eshleman SH, Hackett J Jr, Swanson P, Cunningham SP, Drews B, Brennan C, Devare SG, Zekeng L, Kaptue L, Marlowe N (2004) Performance of the Celera Diagnostics ViroSeq™ HIV-1 genotyping system for sequence-based analysis of diverse human immunodeficiency virus type 1 strains. J Clin Microbiol 42:2711–2717
Ribas SG, Leo H, Pascale O, Katrien F (2006) Performance evaluation of the two protease sequencing primers of the Trugene HIV-1 genotyping kit. J Virol Methods 135:137–142
Clavel F, Hance AJ (2004) HIV drug resistance. N Engl J Med 350:1023–1035
Engelbrecht S, VanZyl G, Claassen M, Laten A, Jacobs G, Preiser W (2007) Sensitive in-house RT-PCR assay for the assessment of HIV-1 antiretroviral drug resistance. SA-AIDS, p 528
Saravanan S, Vidyaa M, Balakrishanana P, Kumarasamya N, Solomona SS, Solomona S, Kantor R, Katzensteinc D, Ramratnamb B, Mayer KH (2009) Evaluation of two human immunodeficiency virus-1 genotyping systems: ViroSeq™ 20 and an in-house method. J Virol Methods 159:211–216
Wallis CL, Papathanasopoulos MA, Lakhi S, Karita E, Kamali A, Kaleebu P, Sanders E, Anzala O, Bekker LG, Stevens G, de Wit TF, Stevens W (2010) Affordable in-house antiretroviral drug resistance assay with good performance in non-subtype B HIV-1. J Virol Methods 163:505–508
Lee S, Espin F, Turner J, Griswold M, Kahn D (1999) Genotyping of HIV-1 drug-resistance: comparison between in-house procedure and visible genetics Trugene [Tm] HIV-1 assay. Abstr Intersci Conf Antimicrob Agents Chemother 39:462
Loubsher S, Sherman G, Chezzi C, Jones J, Cohen S, Puren A, Morris L (2004) Characterization of nevirapine resistance mutations using RT-PCR and DNA sequencing methods in a mother-infant cohort following single dose nevirapine. Antivir Ther 9:S145
Steegen K, Demecheleer E, De Cabooter N, Nges D, Temmerman M, Ndumbe P, Mandaliya K, Plumb J, Verhofstede C (2006) A sensitive in-house RT-PCR genotyping system for combined detection of plasma HIV-1 and assessment of drug resistance. J Virol Methods 133:137–145
Lindstrom A, Albert J (2003) A simple and sensitive in-house method for determining genotypic drug resistance of HIV-1. J Virol Methods 107:45–51
Laurent C, Kouanfack C, Vergne L, Tardy M, Zekeng L, Noumsi N, Butel C, Bourgeois A, Mpoudi-Ngole E, Koulla-Shiro S, Peeters M, Delaporte E (2006) Antiretroviral drug resistance and routine therapy. Cameroon Emerg Infect Dis 12:1001–1004
Aghokeng FA, Vergne L, Mpoudi NE, Mbangue M, Deoudje N, Mokondji E, Nambei WS, Peyou-Ndi MM, Moka JJL, Delaporte E, Peeters M (2009) Evaluation of transmitted HIV drug resistance among recently-infected antenatal clinic attendees in four Central African countries. Antiviral Therapy 14:401–411
Kouanfack C, Montavon C, Laurent C, Aghokeng A, Kenfack A, Bourgeois A, Koulla-Shiro S, Mpoudi-Ngole E, Peeters M, Delaporte E (2009) Low levels of antiretroviral-resistant HIV infection in a routine clinic in Cameroon that uses the World Health Organization (WHO) public health approach to monitor antiretroviral treatment and adequacy with the WHO recommendation for second-line treatment. Clin Infect Dis 48:1318–1322
Hall T (2001) BioEdit version 506. Department of Microbiology, North Carolina State University, copyright 1997–2001. Last updated on 2/12/2001
Swofford DL (2002) PAUP phylogenetic analysis using parsimony (*and other methods): version 40. Sinauer Associates, Sunderland
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol 53:793–808
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Kosakovsky PSL, Posada D, Stawiski E, Chappey C, Poon AF, Hughes G, Fearnhill E, Gravenor MB, Leig BAJ, Frost SD (2009) An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comput Biol 5(11):e1000581
Struck D, Perez BD, Devaux C, Schmit JC (2010) COMET: a novel approach to HIV-1 subtype prediction. 8th European HIV drug resistance workshop. Abstract 88
Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC (1999) Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160
Maes B, Schrooten Y, Snoeck J, Derdelinckx I, Van Ranst M, Vandamme AM, van Laethem K (2004) Performance of ViroSeq HIV-1 genotyping system in routine practice at a Belgian clinical laboratory. J Virol Methods 119:45–49
Diana DH, Susan HE, Donald JB, Paul EP, James WB (2003) Evaluation of the editing process in human immunodeficiency virus type 1 genotyping. J Clin Microbiol 41:3265–3272
Peeters M, Toure-Kane C, Nkengasong JN (2003) Genetic diversity of HIV in Africa: impact on diagnosis, treatment, vaccine development and trials. AIDS 17:2547–2560
Butler IF, Pandrea I, Marx PA, Apetrei C (2007) HIV genetic diversity: biological and public health consequence. Curr HIV Res 5:23–45
Mitchell C, Kraft K, Peterson D, Frenkel L (2010) Cross-contamination during processing of dried blood spots used for rapid diagnosis of HIV-1 infection of infants is rare and avoidable. J Virol Methods 163:489–491
Duchassaing D, Dingeon B, Chalas J, Coude M, Coulhon MP, Launay JM (2002) Molecular genetics: pre- and post-analytic “best practices”. Ann Biol Clin (Paris) 60:617–621
Hamers RL, Derdelinckx I, van Vugt M, Stevens W, Rinke de Wit TF, Schuurman R (2008) The status of HIV-1 resistance to antiretroviral drugs in sub-Saharan Africa. Antivir Ther 13:625–639
Acknowledgments
This work was supported financially by grants from the Italian National Institute of Health, Rome, through funding from the Italian Cooperation to the government of Cameroon, and by the European Commission Framework 7 Programme (CHAIN, the Collaborative HIV and Anti-HIV Drug Resistance Network, Integrated Project no. 223131). We would like to thank Prof Lutz Gürtler for his precious contributions in the revision of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
J. Fokam and R. Salpini contributed equally to this work.
Rights and permissions
About this article
Cite this article
Fokam, J., Salpini, R., Santoro, M.M. et al. Performance evaluation of an in-house human immunodeficiency virus type-1 protease-reverse transcriptase genotyping assay in Cameroon. Arch Virol 156, 1235–1243 (2011). https://doi.org/10.1007/s00705-011-0982-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-011-0982-3