Abstract
The family Totiviridae includes a number of viruses with monosegmented dsRNA genomes and isometric virions that infect either fungi or a number of medically important protozoan parasites such as Leishmania and Giardia. A new genus, Trichomonasvirus, was recently approved for this family. Its name is based on the genus of its host organism, Trichomonas vaginalis, a protozoan parasite that colonizes the human genitourinary mucosa and is the most common non-viral sexually transmitted infection in the world. The type species of this new genus is Trichomonas vaginalis virus 1. Distinguishing characteristics of the new genus include infection of a human sexually transmitted parasite, stable mixed infection with more than one distinct Trichomonasvirus species, and sequence-based phylogenetic divergence that distinguishes it from all other family members.
Similar content being viewed by others
Introduction
Members of the family Totiviridae are characterized by icosahedral virions, ranging between 30 and 40 nm in diameter, which normally encapsidate monosegmented (i.e., nonsegmented) double-stranded RNA (dsRNA) genomes with overlapping open reading frames encoding a capsid protein (CP) and an RNA-dependent RNA polymerase (RdRp) [26]. In some members of this family (e.g., those from the genera Giardiavirus, Leishmaniavirus, and Totivirus), the RdRp is expressed as a CP/RdRp fusion protein, either as a consequence of ribosomal frameshifting [15, 32] or as a direct fusion with CP [34]. In other members (e.g., those from the genus Victorivirus), it is expressed as a separate, nonfused protein [31].
The greater than 25 species in the family Totiviridae that have been recognized by the International Committee on Taxonomy of Viruses (ICTV) as of spring 2009 are known to mediate largely noncytopathic, persistent infections of a diverse range of fungi and protozoa. Four genera are currently recognized: Giardiavirus, Leishmaniavirus, Totivirus, and Victorivirus [26]. Viruses that infect protozoa have been assigned to the former two genera, and those that infect fungi to the latter two.
Trichomonas vaginalis is a sexually transmitted, flagellated protozoan that colonizes the mucosal epithelium of the human genitourinary tract, most consistently the vagina in women and urethra in men, causing inflammatory disease and associated risks of reproductive problems [17]. Trichomoniasis is the most common curable sexually transmitted disease in the world, with estimated prevalence higher than that of syphilis, gonorrhoea, and chlamydia combined [24]. The prevalence of trichomoniasis in inner-city, sexually transmitted infection (STI) clinics in the USA approaches 25%, and trichomoniasis is estimated to account for almost 50% of all curable infections worldwide [47], or an estimated 170 million new cases each year [63]. In addition to the morbidity associated with symptomatic vaginitis (pruritus, irritation, and discharge) and/or urethritis, it has also been associated with a variety of other clinical outcomes, including low birth weight and premature delivery [13], and an increased risk of transmission and acquisition of human immunodeficiency virus, human papillomavirus, and cervical cancer [40, 64]. Recurrence of T. vaginalis infection, reflecting lack of a protective immune response in the human host, and resistance to the drug of choice metronidazole are other associated problems [47]. Virulence of the parasite is mediated by several factors including cysteine proteases, surface proteins, and the major surface lipophosphoglycan, the latter being responsible for selective upregulation of inflammatory mediators by the host cervical and vaginal epithelial cells [18]. Immunosuppressive effects on bystander immune cells have also been observed in vitro; however, their molecular basis remains poorly understood [17].
The presence of long, linear dsRNA molecules in many strains of T. vaginalis was first reported in 1985, followed shortly by evidence of its association with virus-like particles [19, 53, 54]. Further screening of T. vaginalis strains revealed that the presence of viral dsRNA was relatively common, and that as many as three dsRNA segments of similar length (4,000–5,000 bp) could be identified in a single isolate, suggesting either the presence of a multisegmented virus or the possibility of mixed infection by several different Trichomonas vaginalis viruses (TVVs) [20, 35]. Genome sequencing and phylogenetic analyses of several TVV genomes (Table 1) subsequently demonstrated homology to monosegmented dsRNA viruses of the family Totiviridae [10, 49, 51]. Based on relatedness of the Trichomonas and Giardia host organisms, TVVs were tentatively assigned to the genus Giardiavirus, of which Giardia lamblia virus is the well-characterized type species [55, 57]. Further genome sequencing and phylogenetic analyses, however, have suggested that TVVs are not so closely related to G. lamblia virus (GLV) as to warrant grouping in the same genus [26, 39].
The phylogenetic divergence and other distinguishing characteristics of TVVs recently led to the proposal of a new genus in the family Totiviridae to accommodate them. The establishment of this new genus, named Trichomonasvirus based on the host organism and in parallel with the genus names for other protozoan viruses in the family (Giardiavirus and Leishmaniavirus), has been recently approved by the Executive Committee of the ICTV and is awaiting ratification at the next ICTV meeting, to be held in Sapporo, Japan, in September 2011.
Taxonomic structure
- Order::
-
Unassigned
- Family::
-
Totiviridae
- Genus::
-
Trichomonasvirus
- Type species::
-
Trichomonas vaginalis virus 1
- Other species::
-
Trichomonas vaginalis virus 2
- :
-
Trichomonas vaginalis virus 3
TVVs were first tentatively assigned to the genus Giardiavirus given that both hosts (T. vaginalis and G. lamblia) are related flagellates. However, these protozoa are generally now assigned to separate major clades—Parabasalia (order Trichomonadida) and Fornicata (order Diplomonadida), respectively—within the supergroup Excavata [1], and recent genome sequencing studies have confirmed that T. vaginalis and G. lamblia are indeed well differentiated at the genomic level [11, 43]. Moreover, genome sequences of their respective viruses have revealed that TVVs are phylogenetically divergent from GLV [26, 39]. These findings, along with phylogenetic clustering of the three proposed species of TVVs, justify their assignment to a new genus in the family Totiviridae, apart from genus Giardiavirus.
Trichomonas vaginalis virus 1 was chosen as the type species for the new genus because its prototypical strain Trichomonas vaginalis virus 1-1 (TVV1-1) was the first virus for which full-length genome sequence data was reported [51] and because TVV1-1 and other TVV1 strains remain the best characterized and most commonly reported to date. Of the currently five full-length genomic sequences for TVVs in GenBank, three are from different strains of TVV1. The remaining two are from Trichomonas vaginalis virus 2-1 (TVV2-1) and Trichomonas vaginalis virus 3-1 (TVV3-1), representing the protoypical strains of the two other newly proposed species, Trichomonas vaginalis virus 2 [10] and Trichomonas vaginalis virus 3 [9], respectively (Tables 1, 2). In addition, full-length protein-coding sequences have been reported for a fourth strain of TVV1 [65] (Table 2).
An amino acid (aa) sequence identity of <50% was previously adopted as a cut-off for identifying distinct species in the family Totiviridae [62]. By this criterion, pairwise comparison of the six TVV isolates with full-length protein-coding sequences justifies the proposal of three distinct TVV species, with interspecies aa sequence identities of ≤32 and ≤43% for CP and RdRp, respectively (Table 2). Reciprocally, the four reported strains of TVV1 have ≥83% aa sequence identity in pairwise comparisons of the two proteins (Table 2).
Biological properties
Like most other members of the family Totiviridae, TVVs have not been shown capable of extracellular transmission and likely lack the molecular machinery for either exit or entry of their protozoan host. Rather, they are transmitted vertically during mitotic cell division, that is, during the asexual reproduction of T. vaginalis trophozoites by binary fission [47]. Transmission of TVVs during meiotic cell division and then by cell–cell fusion upon mating might also occur. Although mating by T. vaginalis has not yet been observed, evidence for its occurrence includes (1) chromosomal synapses typical of meiosis in a small subset of cultured T. vaginalis cells [16], (2) genetic variation among T. vaginalis strains sufficient to suggest meiotic recombination [30], and (3) the presence of a number of proteins uniquely required for meiosis encoded in the T. vaginalis genome [11, 42]. T. vaginalis does not form cysts, and transmission between human hosts is instead generally by transfer of trophozoites during sexual contact [47].
TVVs are considered to be well adapted to T. vaginalis such that they cause few if any deleterious effects in the protozoan and maintain a largely noncytopathic, persistent infection. In our own experience, TVVs can remain stably associated with the T. vaginalis host for 6 months or more of serial passages in vitro with little or no changes in the parasite proliferative cycle (RN Fichorova and ML Nibert, unpublished data). The literature, however, reports occasional examples of TVV-associated cytopathology in T. vaginalis [8, 12, 46]. Thus, there may be particular circumstances in which the détente achieved by this virus-host pair is broken.
Despite the usual lack of overt harm to T. vaginalis, TVV infection has been demonstrated to alter cell-surface expression of a highly immunogenic T. vaginalis protein, P270 [2]. T. vaginalis strains infected with the virus are heterogeneous with respect to the presence of the surface immunogen and display phenotypic variation upon cultivation, whereas those without virus infection display a stable negative phenotype [56]. In one study, expression of the surface immunogen in the presence of TVV infection was increased at the mRNA level [36], and in another study, upregulation of P270 resulted in diminished ability to cause contact-dependent cytotoxicity of HeLa cells [2]. In a crude mouse model of pathogenicity, which has limited physiologic relevance to the natural mucosal infection, subcutaneously injected TVV-infected T. vaginalis demonstrated that one freshly isolated TVV-positive strain caused weaker abscess formation relative to seven TVV-negative and two other TVV-positive T. vaginalis strains [39]. Collectively, these sparse data suggest that TVV infection may impair the clearance of the T. vaginalis infection and allow its longer persistence in the host. Further basic research is needed to elucidate all aspects of virus–parasite–human interactions.
TVV infection has also been shown to alter cysteine proteinase expression profiles in T. vaginalis [46]. Cysteine proteinases are known virulence factors of T. vaginalis, responsible for degradation of a number of key proteins in vaginal immune defenses, such as proteins of the complement cascade [3] and immunoglobulins [45], and also facilitate cytoadherence [6]. It is thus possible that TVV modulation of T. vaginalis proteinase expression may in turn increase its survival in the human host. Once again, more studies are needed to correlate TVV infection with T. vaginalis pathogenicity and clinical outcome.
Much remains unknown about the specifics of the cell biology of TVV infections, including the subcellular localization of their replication and assembly complexes. An association of some virus-like particles with the Golgi apparatus in TVV-infected T. vaginalis has been described [8], but the significance of this observation remains unclear.
The extent to which related dsRNA viruses may infect other trichomonads is unclear. A recent study identified VLPs in isolates of Tritrichomonas foetus, a bovine pathogen, but required treatment with various cytoskeleton-affecting drugs to visualize the VLPs through electron microscopy [29]. On the other hand, a recent examination of isolates of Trichomonas gallinae, a significant fowl pathogen, revealed no evidence of viruses by electron microscopy or RNA purification [25] but did not include similar drug treatments as in the Tritrichomonas foetus study.
Virions and replication
TVVs have isometric virions between 30 and 40 nm in diameter, as visualized by negative staining and transmission electron microscopy [8, 10, 54]. The capsids appear single layered and thin, without obvious protrusions, and have buoyant densities between 1.33 and 1.39 g/cm3 on CsCl gradients. Higher-resolution structures remain to be determined.
From genome-based predicted protein sequences, the CPs of different TVVs are similar in size, ranging from 678 to 709 aa in length (Table 1), or roughly 75 to 79 kDa in molecular mass. These values are similar to those of the CPs of several other members of the family Totiviridae [26]. The RdRp open reading frames (ORFs) are also similar in size among different TVVs, ranging from 756 to 767 aa in length (Table 1), or roughly 86 to 88 kDa in molecular mass. These values are similar to those of the CP and RdRp ORFs of several other members of the family Totiviridae [26]. Given the overlapping ORFs of the CP and RdRp in TVVs, the RdRp in each is believed to be expressed as a fusion protein with the CP (see below).
The genome of each TVV is a single dsRNA molecule, approximately 4,600 to 4,900 bp in length. It is unknown at this time whether the genome is 5′-capped at either end. As many as three dsRNA molecules of roughly similar size may be isolated from a single strain of T. vaginalis in association with purified virus particles [35]; however, sequencing information from these molecules suggests co-infection with distinct TVVs, rather than multisegmented viruses. In addition, smaller dsRNA molecules ~500 to ~1,700 bp in length may be isolated from some strains of T. vaginalis infected with TVVs, which likely represent satellite dsRNAs [10, 37, 38, 50] (see below).
RNA transcription by TVVs is expected to be like that by other members of the family Totiviridae: asymmetric (producing only plus-strand transcripts), end-to-end (producing full-length plus-strand transcripts that mimic the genomic plus strand), and conservative (retaining both parental strands, plus and minus, within the transcribing virus particles) [23, 27, 44, 59]. A single round of minus-strand synthesis using a full-length plus-strand RNA as template is thought to generate the duplex RNA genome in newly assembling virions.
Genomic and coding properties
Full-length nucleotide sequences of five TVV strains representing the three distinct species have been deposited in GenBank (Table 1). Each virus has a dsRNA genome ranging in length between 4,647 and 4,844 bp.
The genome plus strand of each TVV encodes at least two long ORFs (Fig. 1). The upstream ORF (ORF1) encodes the CP, whereas the downstream ORF (ORF2) encodes the RdRp [9, 10, 39, 49, 51, 65]. The two ORFs are in different reading frames, which overlap by 16–123 nt. The RdRp of each virus is thereby thought to be expressed as a CP/RdRp fusion protein following either a +1 (TVV1) or −1 (TVV2, TVV3) ribosomal frameshift. Putative slippery heptanucleotides for promoting ribosomal frameshifting have been identified (TVV1, TVV2) [10, 39, 49, 51, 65] or can be predicted (TVV3) in the overlapping segments of the CP and RdRp ORFs (Fig. 2). The previous reports of TVV sequences have failed to identify RNA pseudoknots or other RNA structures that have been commonly associated with ribosomal frameshifting sites in other viruses, etc. [28]. Expression of a 160,000-Mr CP/RdRp fusion protein (sequence-predicted molecular mass 160 and 162 kDa, respectively) has been shown by western blotting for both TVV1 and TVV2 [10, 41].
Coding diagrams for TVV1, TVV2, and TVV3. Open reading frames (ORFs) 1 [capsid protein (CP, red in online version of figure)] and 2 [RNA-dependent RNA polymerase (RdRp), blue in online version of figure] are diagrammed for each virus. The first and last codons for each ORF are indicated; in the case of ORF1, the first codon is considered to be that of the proposed AUG start codon. In each virus, the RdRp is thought to be expressed as a CP/RdRp fusion following ribosomal frameshifting as indicated. 5′ and 3′ untranslated regions (UTRs) are also labeled, along with the position numbers of the first and last nucleotides of each genome and ORF. The specific nucleotides and nucleotide position numbers shown in this figure as well as in Fig. 2 are for the prototypical strains TVV1-1 [51], TVV2-1 [10], and TVV3-1 [9]
Regions of ribosomal frameshifting in TVV1, TVV2, and TVV3. Virus is labeled at the top of each sequence block, along with the position numbers of the displayed nucleotides from the genomic plus-strand sequence. The nature of the putative frameshifting event in each is also shown at top. The putative slippery heptanucleotide involved in signalling the ribosomal frameshift event in each virus is underlined in its genomic sequence. For each virus, beneath each of the three displayed lines of genomic sequence, the amino acid translation for all three frames is shown. Any stop codons in each frame are shown as filled circles. The genomic sequences were chosen for display such that the long ORF for CP (red in online version of figure) appears in the middle frame. In TVV1, the long ORF for RdRp (blue in online version of figure) therefore appears in the bottom frame (+1 frameshift), whereas in TVV2 and TVV3, it appears in the top frame (−1 frameshift)
In the single reported TVV2 genome sequence, third and fourth long ORFs (encoding >200 aa each) have been noted in a central portion of the remaining reading frame [10]. The putative proteins encoded by these ORFs are very basic (pI = 11) and rich in Ser and Thr residues, with no significant similarity to protein sequences in GenBank, and there is no gel evidence for their expression [10]. Among the reported TVV1 and TVV3 sequences, additional ORFs of similarly long length are not present in the remaining reading frame, although one or two ORFs of >100 aa each can be found in each strain. The significance of any of these additional ORFs in TVV1, TVV2, and TVV3 remains open to question. The genomic minus strands of the TVVs similarly contain only short ORFs in all three reading frames, consistent with their expected lack of availability for translation due to sequestration in viral capsids throughout the replication cycle.
The 5′ and 3′ untranslated regions (UTRs) of TVVs range from 285 to 359 nt and 69 to 154 nt, respectively, and demonstrate little sequence identity between species. Secondary structure predictions of the 5′ and 3′ ends of the plus strands of TVV1 have suggested the presence of a large 3′ stem-loop structure, but no analogous 5′ stem loop [49, 51]. Similar analyses for TVV2 have revealed stem-loop structures at both ends of the plus strand [10]. Such RNA secondary structures may act as signals for RNA replication and/or packaging.
Some isolates of T. vaginalis infected with TVVs are also associated with smaller dsRNA molecules, ranging in size from ~500 to ~1,700 bp [10, 37, 38, 50]. Experiments suggest that these dsRNAs can be packed into TVV capsids independently from genomic dsRNA [50], can serve as specific templates for the viral RdRp [38], do not encode their own RdRp [37, 50], and have little sequence homology with the TVV genome [37], consistent with satellite dsRNAs. Sequencing of several satellites has revealed them to have only short ORFs in all three reading frames of both strands, suggesting a lack of protein-coding ability [37, 50]. While the biological significance of the satellite dsRNAs associated with TVVs is unclear, dsRNA satellites are commonly associated with fungal viruses in the family Totiviridae [26, 61, 62].
Phylogenetic relationships
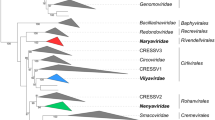
The full-length protein-coding sequences for six TVV strains that have been deposited into GenBank (Table 2) allow phylogenetic comparisons among these viruses as well as with other members of the family Totiviridae. From such analyses, it is clear that TVVs constitute a monophyletic cluster distinguishable from all other viruses in the family (Fig. 3), including from members of the genus Giardiavirus, to which TVVs had been previously assigned on a tentative basis before the recently approved proposal to segregate them into their own new genus, Trichomonasvirus. Moreover, the TVVs appear to be more closely related to protozoan viruses in the genus Leishmaniavirus and to fungal viruses in the genus Victorivirus than to other protozoan and fungal viruses in the respective genera Giardiavirus and Totivirus.
Phylogenetic relationships among TVVs and other approved members of the family Totiviridae. Maximum-likelihood trees were derived from the full-length, concatenated CP and RdRp ORF sequences of each analyzed virus. The multiple sequence alignment was generated using MUSCLE v3.7 without subsequent Gblocks curation; the phylogenetic tree was generated using PhyML v3.0; the confidence index for each branch (expressed as % in the labels) was determined as described by Anisimova and Gascuel [5]; and the tree was rendered using TreeDyn v198.3. All four steps were performed using the “A la Carte” option at http://www.phylogeny.fr/ [14]. The tree was additionally refined for presentation using FigTree v1.3.1 obtained from http://www.tree.bio.ed.ac.uk/software/figtree/. The tree is unrooted, and the scale bar indicates the number of substitutions per position in the alignment. See text for the GenBank accession no. of each TVV sequence (prototypical strains TVV1-1, TVV2-1, and TVV3-1). Other members of the family Totiviridae included in the tree are (GenBank Accession No. in parenthesis after each): LRV1-1, Leishmania RNA virus 1-1 (M92355); LRV2-1, Leishmania RNA virus 2-1 (U32108), LRV1-4, Leishmania RNA virus 1-4 (U01899); HvV190S, Helminthosporium victoriae virus 190S (HVU41345); CeRV1, Chalara elegans RNA virus 1 (AY561500); HmV1-17, Helicobasidium mompa totivirus 1-17 (AB085814); MoV1, Magnaporthe oryzae virus 1 (AB176964); SsRV1, Sphaeropsis sapinea RNA virus 1 (AF038665); CmRV, Coniothyrium minitans RNA virus (AF527633); EfV1, Epichloe festucae virus 1 (AM261427); GaRV-L1, Gremmeniella abietina RNA virus L1 (AF337175); SsRV2, Sphaeropsis sapinea RNA virus 2 (AF039080); ScV-LA, Saccharomyces cerevisiae virus LA (J04692); ScV-LBC, Saccharomyces cerevisiae virus LBC (U01060); UmV-H1, Ustilago maydis virus H1 (U01059); and GLV (L13218). Viruses are clustered and labeled as follows: genus Trichomonasvirus (magenta in online version), genus Leishmaniavirus (green in online version), genus Victorivirus (orange in online version), genus Totivirus (cyan in online version), and genus Giardiavirus (brown in online version)
Among TVVs, TVV2 and TVV3 strains appear more closely related to each other than either is to TVV1 strains (Fig. 3; also see Table 2). This parallels a basic difference in coding strategy in that all four reported strains of TVV1 express their RdRps following a +1 ribosomal frameshift, whereas both TVV2 and TVV3 strains do so following a −1 frameshift. The TVV1 strains also seem to lack a predicted stem-loop structure at their 5′ termini that the TVV2 and TVV3 strains contain. The four TVV1 strains show limited divergence (see Table 2), and sequences of additional TVV2 and TVV3 strains are needed to determine if they each also constitute such a well-delimited group.
Clinical relevance
T. vaginalis is the most common nonviral STI in the world and is the causal agent of trichomoniasis [63]. The severity of morbidity varies broadly, from asymptomatic to severely symptomatic vaginitis, and complications affect nearly every aspect of reproductive health including risks of pelvic inflammatory disease and tubal infertility, premature membrane rupture, preterm delivery, low birth weight, post-hysterectomy cuff cellulitis, and viral STIs including human immunodeficiency virus and human papillomavirus [17]. Given the widespread prevalence of TVV infections (see below) and the demonstration that TVV infections can modulate host translation and expression of immunogenic proteins [2], it is possible that underlying TVV infections may modulate the pathogenesis of T. vaginalis infections and possibly underlie differences in symptomatology, drug resistance, parasite load, transmissibility, etc. There is precedence in the literature for endobiotic viruses modulating the pathogenicity or behavior of their parasitic hosts; for example, a virus infecting Cryptosporidium parvum has been shown to increase the fecundity of its protozoan host (levels of oocysts shed in feces) during C. parvum infection of dairy calves [33].
While a few reports have suggested that T. vaginalis infection with TVV may result in attenuated cytopathogenicity and acute inflammatory response [39], others suggest the possibility of hypervirulence through upregulation of a major surface immunogen [2] and upregulation of host proteinase expression [46]. A study of adolescents with T. vaginalis infection and no other STIs showed that the presence of vaginal discharge, dysuria, dyspareunia, and cervical erythema, but not pruritus, vulvar or vaginal erythema, were significantly associated with TVV infection diagnosed by gel electrophoresis of nucleic acids, suggesting its clinical relevance [22]. More studies are needed to correlate TVV-infected T. vaginalis with symptomatology or pathogenicity in infected humans.
The prevalence of TVV in different T. vaginalis clinical isolates varies. Rates as high as 82 and 75% were found in clinical isolates from South Africa [58] and Baltimore [60], respectively, whereas rates closer to 50% were noted in isolates from Cuba (55%) [21], the USA (50%) [48], and various geographic locations (44%) [52]. In each of the listed studies, TVVs were detected by gel electrophoresis of nucleic acids. Opportunities for improved diagnostics of TVV infection include immunodetection methods with TVV-specific antibodies [4] and nucleic acid microarrays to detect TVV RNAs [7].
References
Adl SM, Simpson AG, Farmer MA et al (2005) The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol 52:399–451
Alderete JF, Kasmala L, Metcalfe E, Garza GE (1986) Phenotypic variation and diversity among Trichomonas vaginalis and correlation of phenotype with trichomonal virulence determinants. Infect Immun 53:285–293
Alderete JF, Provenzano D, Lehker MW (1995) Trichomonas vaginalis resistance to complement is mediated by iron-inducibile cysteine poteinase. Microbiol Patol 19:93–103
Alderete JF, Wendel KA, Rompalo AM, Erbelding EJ, Benchimol M, Chang TH (2003) Trichomonas vaginalis: evaluating capsid proteins of dsRNA viruses and the dsRNA virus within patients attending a sexually transmitted disease clinic. Exp Parasitol 103:4450
Anisimova M, Gascuel O (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55:539–552
Arroyo R, Alderete JF (1989) Trichomonas vaginalis proteinase activitiy is necessary for parasite adherence to epithelial cells. Infect Immun 57:2991–2997
Baptista CS, Wu X, Munroe DJ (2007) International patent WO/2007/130519, Viral nucleic acid microarray and method of use. World Intellectual Property Organization, Geneva, Switzerland
Benchimol M, Monteiro S, Chang TH, Alderete JF (2002) Virus in Trichomonas—an ultrastructural study. Parasitol Int 51:293–298
Bessarab IN, Tai JH (2001) Trichomonas vaginalis virus 3 capsid protein (cap) and RNA-dependent RNA polymerase (pol) genes, complete cds. Genbank, Accession No. AF325840
Bessarab IN, Liu HW, Ip CF, Tai JH (2000) The complete cDNA sequence of a type II Trichomonas vaginalis virus. Virology 267:350–359
Carlton JM, Hirt RP, Silva JC et al (2007) Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315:207–212
Champney WS, Curtis SK, Samuels R (1995) Cytopathology and release of an RNA virus from a strain of Trichomonas vaginalis. Int J Parasitol 25:1463–1471
Cotch MF, Pastorek JG 2nd, Nugent RP et al (1997) Trichomonas vaginalis associated with low birth weight and preterm delivery. The Vaginal infections and Prematurity Study Group. Sex Transm Dis 24:1–8
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469
Diamond ME, Dowhanick JJ, Nemerff ME, Pietras DF, Tu C, Bruenn JA (1989) Overlapping genes in a yeast double-stranded RNA virus. J Virol 63:3983–3990
Drmota T, Král J (1997) Karyotype of Trichomonas vaginalis. Eur J Protistol 33:131–135
Fichorova RN (2009) Impact of T. vaginalis infection on innate immune responses and reproductive outcome. J Reprod Immunol 83:185–189
Fichorova RN, Trifonova RT, Gilbert RO, Costello CE, Hayes GR, Lucas JJ, Singh BN (2006) Trichomonas vaginalis lipophosphoglycan triggers a selective upregulation of cytokines by human female reproductive tract epithelial cells. Infect Immun 74:5773–5779
Flegr J, Cerkasov J, Kulda J, Tachezy J, Stokrová J (1987) The dsRNA of Trichomonas vaginalis is associated with virus-like particles and does not correlate with metronidazole resistance. Folia Microbiol (Praha) 32:345–348
Flegr J, Cerkasov J, Stokrová J (1988) Multiple populations of double-stranded RNA in two virus-harbouring strains of Trichomonas vaginalis. Folia Microbiol (Praha) 33:462–465
Fraga J, Rojas L, Sariego I, Fernández-Calienes A (2005) Double-stranded RNA viral infection in Cuban Trichomonas vaginalis isolates. Braz J Infect Dis 9:521–524
Fraga J, Rojas L, Sariego I, Fernandez-Calienes A, Nunez FA (2007) Double-stranded RNA viral infection of Trichomonas vaginalis and association with clinical presentation. Acta protozool 46:93–98
Fujimura T, Esteban R, Wickner RB (1986) In vitro L-A double-stranded RNA synthesis in virus-like particles from Saccharomyces cerevisiae. Proc Natl Acad Sci USA 83:4433–4437
Gerbase AC, Rowley JT, Heymann DH, Berkley SF, Piot P (1998) Global prevalence and incidence estimates of selected curable STDs. Sex Transm Infect 74:S12–S16
Gerhold RW, Allison AB, Sellers H, Linnemann E, Chang TH, Alderete JF (2009) Examination for double-stranded RNA viruses in Trichomonas gallinae and identification of a novel sequence of a Trichomonas vaginalis virus. Parasitol Res 105:775–779
Ghabrial SA (2008) Totiviruses. In: Mahy BWJ, Van Regenmortel MHV (eds) Encyclopedia of Virology, vol 5, 3rd edn. Elsevier, San Diego, pp 163–174
Ghabrial SA, Havens WM (1989) Conservative transcription of Helminthosporium victoriae 190S virus dsRNA in vitro. J Gen Virol 70:1025–1035
Giedroc DP, Cornish PV (2009) Frameshifting RNA pseudoknots: structure and mechanism. Virus Res 139:193–208
Gomes Vancini R, Benchimol M (2005) Appearance of virus-like particles in Tritrichomonas foetus after drug treatment. Tissue Cell 37:317–323
Hampl V, Vanácová S, Kulda J, Flegr J (2001) Concordance between genetic relatedness and phenotypic similarities of Trichomonas vaginalis strains. BMC Evol Biol 1:11
Huang S, Ghabrial SA (1996) Organization and expression of the double-stranded RNA genome of Helminthosporium victoriae 190S virus, a totivirus infecting a plant pathogenic filamentous fungus. Proc Natl Acad Sci USA 93:12541–12546
Icho T, Wickner RB (1989) The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J Biol Chem 264:6717–6723
Jenkins MC, Higgins J, Abrahante JE, Kniel KE, O’Brien C, Trout J, Lancto CA, Abrahamsen MS, Fayer R (2008) Fecundity of Cryptosporidium parvum is correlated with intracellular levels of the viral symbiont CPV. Int J Parasitol 38:1051–1055
Kang J, Wu J, Bruenn JA, Park C (2001) The H1 double-stranded RNA genome of Ustilago maydis virus-H1 encodes a polyprotein that contains structural motifs for capsid polypeptide, papain-like protease, and RNA-dependent RNA polymerase. Virus Res 76:183–189
Khoshnan A, Alderete JF (1993) Multiple double-stranded RNA segments are associated with virus particles infecting Trichomonas vaginalis. J Virol 67:6950–6955
Khoshnan A, Alderete JF (1994) Trichomonas vaginalis with a double-stranded RNA virus has upregulated levels of phenotypically variable immunogen mRNA. J Virol 68:4035–4038
Khoshnan A, Alderete JF (1995) Characterization of double-stranded RNA satellites associated with the Trichomonas vaginalis virus. J Virol 69:6892–6897
Khoshnan A, Provenzano D, Alderete JF (1994) Unique double-stranded RNAs associated with the Trichomonas vaginalis virus are synthesized by viral RNA-dependent RNA polymerase. J Virol 68:7108–7114
Kim JW, Chung PR, Hwang MK, Choi EY (2007) Double-stranded RNA virus in Korean isolate IH-2 of Trichomonas vaginalis. J Parasitol 45:87–94
Laga M, Alary M, Nzila N, Manoka AT, Tuliza M, Behets F, Goeman J, St Louis M, Piot P (1994) Condom promotion, sexually transmitted disease treatment, and declining incidence of HIV-1 infection in female Zairian sex workers. Lancet 344:246–248
Liu HW, Chu YD, Tai JH (1998) Characterization of Trichomonas vaginalis virus proteins in the pathogenic protozoan T. vaginalis. Arch Virol 143:963–970
Malik SB, Pightling AW, Stefaniak LM, Schurko AM, Logsdon JM (2007) An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLoS One 3:e2879
Morrison HG, McArthur AG, Gillin FD et al (2007) Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317:1921–1926
Nemeroff ME, Bruenn JA (1986) Conservative replication and transcription of Saccharomyces cerevisiae viral double-stranded RNA in vitro. J Virol 57:754–758
Provenzano D, Alderete JF (1995) Analysis of human immunoglobulin-degrading cysteine proteinases of Trichomonas vaginalis. Infect Immunol 63:3388–3395
Provenzano D, Khoshnan A, Alderete JF (1997) Involvement of dsRNA virus in the protein composition and growth kinetics of host Trichomonas vaginalis. Arch Virol 142:939–952
Schwebke JR, Burgess D (2004) Trichomoniasis. Clin Microbiol Rev 17:794–803
Snipes LJ, Gamard PM, Narcisi EM, Beard CB, Lehmann T, Secor WE (2000) Molecular epidemiology of metronidazole resistance in a population of Trichomonas vaginalis clinical isolates. J Clin Microbiol 38:3004–3009
Su HM, Tai JH (1996) Genomic organization and sequence conservation in type I Trichomonas vaginalis viruses. Virology 222:470–463
Tai JH, Chang SC, Ip CF, Ong SJ (1995) Identification of a satellite double-stranded RNA in the parasitic protozoan Trichomonas vaginalis infected with T. vaginalis virus T1. Virology 208:189–196
Tai JH, Chui-Fun IP (1995) The cDNA sequence of Trichomonas vaginalis virus-T1 double-stranded RNA. Virology 206:773–777
Vanácová S, Tachezy J, Kulda J, Flegr J (1997) Characterization of trichomonad species and strains by PCR fingerprinting. J Eukaryotic Microbiol 144:545–552
Wang AL, Wang CC (1985) A linear double-stranded RNA in Trichomonas vaginalis. J Biol Chem 260:3697–3702
Wang AL, Wang CC (1986) The double-stranded RNA in Trichomonas vaginalis may originate from virus-like particles. Proc Natl Acad Sci USA 83:7956–7960
Wang AL, Wang CC (1991) Viruses of the protozoa. Annu Rev Microbiol 45:251–263
Wang A, Wang CC, Alderete JF (1987) Trichomonas vaginalis phenotypic variation occurs only among trichomonads infected with the double-stranded RNA virus. J Exp Med 166:142–150
Wang AL, Yang HM, Shen KA, Wang CC (1993) Giardiavirus double-stranded RNA genome encodes a capsid polypeptide and a gag-pol-like fusion protein by a translation frameshift. Proc Natl Acad Sci USA 90:8595–8599
Weber B, Mapeka TM, Maahlo MA, Hoosen AA (2003) Double stranded RNA virus in South African Trichomonas vaginalis isolates. J Clin Pathol 56:542–543
Weeks RS, Patterson JL, Stuart K, Widmer G (1992) Transcribing and replicating particles in a double-stranded RNA virus from Leishmania. Mol Biochem Parasitol 52:207–213
Wendel K, Rompalo A, Erbelding E, Chang TH, Alderete JF (2002) Double-stranded RNA viral infection of Trichomonas vaginalis infecting patients attending a sexually transmitted diseases clinic. J Infect Dis 186:558–561
Wickner RB (2006) Viruses and prions of yeast, fungi, and unicellular organisms. In: Knipe DM, Howley PM (eds) Fields Virology, vol 1, 5th edn. Lippincott Williams & Wilkins, Philadelphia, pp 737–768
Wickner RB, Wang CC, Patterson JL (2005) Family Totiviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Eighth report of the International Committee on Taxonomy of Viruses. Elsevier, San Diego, pp 571–580
World Health Organization (2001) An overview of selected curable sexually transmitted disease. In: Global Programme of AIDS. World Health Organization, Geneva, Switzerland, pp 27–29
Zhang ZF, Begg CB (1994) Is Trichomonas vaginalis a cause of cervical neoplasia? Results from a combined analysis of 24 studies. Int J Epidemiol 23:682–690
Zhao YP, Zhang XC, Chen LF, Li JH, Yin JG, Liu Q, Gong PT (2006) Cloning and sequence analysis of a partial gene of Trichomonas vaginalis dsRNA virus. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi 34:389–390
Acknowledgments
Work on TVV in the Fichorova and Nibert Labs has been supported by a Harvard Catalyst Pilot Grant (RNF and MLN) funded through NIH 5UL 1 RR025758-02. In addition, work on T. vaginalis in the Fichorova Lab has been supported by grants NICHD 2R21HD054451 (RNF) and NIAID 5R01AI079085-01A2 (RNF).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Goodman, R.P., Ghabrial, S.A., Fichorova, R.N. et al. Trichomonasvirus: a new genus of protozoan viruses in the family Totiviridae. Arch Virol 156, 171–179 (2011). https://doi.org/10.1007/s00705-010-0832-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-010-0832-8