Abstract
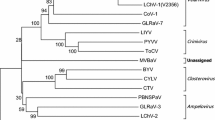
Cherry virus A (CVA) is a graft-transmissible member of the genus Capillovirus that infects different stone fruits. Sweet cherry (Prunus avium L; family Rosaceae) is an important deciduous temperate fruit crop in the Western Himalayan region of India. In order to determine the health status of cherry plantations and the incidence of the virus in India, cherry orchards in the states of Jammu and Kashmir (J&K) and Himachal Pradesh (H.P.) were surveyed during the months of May and September 2009. The incidence of CVA was found to be 28 and 13% from J&K and H.P., respectively, by RT-PCR. In order to characterize the virus at the molecular level, the complete genome was amplified by RT-PCR using specific primers. The amplicon of about 7.4 kb was sequenced and was found to be 7,379 bp long, with sequence specificity to CVA. The genome organization was similar to that of isolates characterized earlier, coding for two ORFs, in which ORF 2 is nested in ORF1. The complete sequence was 81 and 84% similar to that of the type isolate at the nucleotide and amino acid level, respectively, with 5′ and 3′ UTRs of 54 and 299 nucleotides, respectively. This is the first report of the complete nucleotide sequence of cherry virus A infecting sweet cherry in India.


Similar content being viewed by others
References
Eastwell KC, Bernardy MG (1998) Relationship of Cherry virus A to little cherry disease in British Columbia. Acta Hort 472:305–313
Gasic K, Hernandez A, Korban SS (2004) RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Rep 22:437–437
German S, Candresse T, Lanneau M, Huet JC, Pernollet JC, Dunez J (1990) Nucleotide sequence and genomic organization of apple chlorotic leaf spot closterovirus. Virol 179:104–112
Holmes DS, Quigley M (1981) A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem 114:193–197
Isogai M, Aoyagi J, Nakagawa M, Kubodera Y, Sato K, Katoh T, Inamori M, Yamashita K, Yoshikawa N (2004) Molecular detection of five cherry viruses from sweet cherry trees in Japan. J Gen Plant Path 70:288–291
James D, Jelkmann W (1998) Detection of Cherry virus A in Canada and Germany. Acta Hort 472:299–304
James D, Jelkmann W, Upton C (2000) Nucleotide sequence and genome organization of cherry mottle leaf virus and its relationship to members of the Trichovirus genus. Arch Virol 145:995–1007
James D, Varga A, Croft H, Rast H, Thompson D, Hayes S (2006) Molecular characterization, phylogenetic relationships, and specific detection of Peach mosaic virus. Phytopath 96:137–144
Jelkmann W (1994) Nucleotide sequences of apple stem pitting virus and of the coat protein gene of a similar virus from pear associated with vein yellows disease and their relationship with potex- and carlaviruses. J Gen Virol 75:1535–1542
Jelkmann W (1995) Cherry virus A: cDNA cloning of dsRNA, nucleotide sequence analysis and serology reveal a new plant capillovirus in sweet cherry. J Gen Virol 76:2015–2024
Komorowska B, Cieślińska M (2004) First report of Cherry virus A and Little cherry virus-1 in Poland. Plant Dis 88:909
Li R, Mock R (2008) Characterization of a flowering cherry strain of Cherry necrotic rusty mottle virus. Arch Virol 153:973–978
Liberti D, Marais A, Svanella-Dumas L, Dulucq MJ, Alioto D, Ragozzino A, Rodoni B, Candresse T (2005) Characterization of Apricot pseudo-chlorotic leaf spot virus, a novel Trichovirus isolated from stone fruit trees. Phytopath 95:420–426
Mandic B, Matic S, Al Rwahnih M, Jelkmann W, Myrta A (2007) Viruses of sweet and sour cherry in Serbia. J Plant Path 89:103–108
Martelli GP, Adams MJ, Kreuze JF, Dolja VV (2007) Family Flexiviridae: a case study in virion and genome plasticity. Annu Rev Phytopath 45:73–100
Ohira K, Namba S, Rozanov M, Kusumi T, Tsuchizaki T (1995) Complete sequence of an infectious full-length cDNA clone of citrus tatter leaf capillovirus: comparative sequence analysis of capillovirus genomes. J Gen Virol 76:2305–2309
Rao WL, Zhang ZK, Li R (2009) First report of cherry virus A in sweet cherry trees in China. Plant Dis 93:425
Rott ME, Jelkmann W (2001) Complete nucleotide sequence of Cherry necrotic rusty mottle virus. Arch Virol 146:395–401
Sabanadzovic S, Abou Ghanem-Sabanadzovic N, Rowhani A, Grant JA, Uyemoto JK (2005) Detection of Cherry virus A, Cherry necrotic rusty mottle virus and Little cherry virus-1 in California orchards. J Plant Path 87:173–177
Tatineni S, Afunian MR, Gowda S, Hilf ME, Bar-Joseph M, Dawson WO (2009) Characterization of the 50- and 30-terminal subgenomic RNAs produced by a capillovirus: evidence for a CP subgenomic RNA. Virol 385:521–528
Yoshikawa N, Takahashi T (1992) Evidence for translation of Apple stem grooving capillovirus genomic RNA. J Gen Virol 73:1313–1315
Yoshikawa N, Imaizumi M, Takahashi InouyeN (1993) Striking similarities between the nucleotide sequence and genome organization of citrus tatter leaf and apple stem grooving capilloviruses. J Gen Virol 74:2743–2747
Yoshikawa N, Sasaki E, Kato M, Takahashi T (1992) The nucleotide sequence of Apple stem grooving capillovirus genome. Virol 191:98–105
Zhang YP, Kirkpatrick BC, Smart CD, Uyemoto JK (1998) cDNA cloning and molecular characterization of cherry green ring mottle virus. J Gen Virol 79:2275–2281
Acknowledgments
The authors are thankful to the Director, Institute of Himalayan Bioresource Technology (CSIR), Palampur, H.P (India), for providing the necessary facilities. Council of Scientific and Industrial Research, Govt. of India and Department of Biotechnology (DBT), Government of India, are duly acknowledged for financial support (BT/PR/6860/PBD/16/630/2005). Thanks are due to Digvijay Singh Naruka for expert technical assistance with sequencing (IHBT communication number 2080).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Noorani, M.S., Awasthi, P., Singh, R.M. et al. Complete nucleotide sequence of cherry virus A (CVA) infecting sweet cherry in India. Arch Virol 155, 2079–2082 (2010). https://doi.org/10.1007/s00705-010-0826-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-010-0826-6