Abstract
Bats have been identified as a natural reservoir for an increasing number of emerging zoonotic viruses, such as Hendra virus, Nipah virus, Ebola virus, Marburg virus, rabies and other lyssaviruses. Recently, a large number of viruses closely related to members of the genus Coronavirus have been associated with severe acute respiratory syndrome (SARS) and detected in bat species. In this study, samples were collected from 106 live bats of seven different bat species from 27 different locations in Slovenia. Coronaviruses were detected by RT-PCR in 14 out of 36 horseshoe bat (Rhinolophus hipposideros) fecal samples, with 38.8% virus prevalence. Sequence analysis of a 405-nucleotide region of the highly conserved RNA polymerase gene (pol) showed that all coronaviruses detected in this study are genetically closely related, with 99.5–100% nucleotide identity, and belong to group 2 of the coronaviruses. The most closely related virus sequence in GenBank was SARS bat isolate Rp3/2004 (DQ071615) within the SARS-like CoV cluster, sharing 85% nucleotide identity and 95.6% amino acid identity. The potential risk of a new group of bat coronaviruses as a reservoir for human infections is highly suspected, and further molecular epidemiologic studies of these bat coronaviruses are needed.
Similar content being viewed by others
Introduction
The majority of coronaviruses are disease-causing agents [34]. Human coronaviruses are associated with respiratory and gastrointestinal diseases, but coronaviruses detected in animals cause severe respiratory, enteric, neurological or hepatic diseases in their hosts. Coronaviruses were responsible for severe acute respiratory syndrome (SARS), which represents the twenty-first century’s first previously unknown pandemic transmissible disease. SARS coronaviruses isolated from this pandemic are genetically distinct from previously known coronaviruses of animals and humans [39]. SARS coronaviruses have also been isolated from small mammals such as civets (Paguma larvata) and raccoon dogs (Nyctereutes procynoides) from live-animal markets in the southern part of the People’s Republic of China, suggesting that these animals could be the direct source of the SARS epidemic in 2003 [19]. The identification of SARS-like coronaviruses in bats and other wild animals at live-animal markets suggest that a novel human pathogen emerged as a result of interspecies transmission [19]. These findings highlight the potential human-health risk posed by coronaviruses in wild animals. To predict the risk of coronavirus host transition and disease outbreaks, we need to have a deeper understanding of coronavirus reservoirs, such as bats.
Coronaviruses belong to the genus Coronavirus within the family Coronaviridae, order Nidovirales. Coronaviruses are enveloped viruses with positive-stranded RNA genomes of 26–32 kb, the largest continuous RNA genomes in nature [5, 16]. Coronaviruses have a unique mechanism of viral replication that results in a high frequency of recombination and high mutation rates that allow them to adapt to new hosts and niches [26]. On the basis of genetic analysis, coronaviruses are subdivided into three distinct groups: groups 1 and 2 include viruses that are pathogenic for mammals, and group 3 includes viruses that are pathogenic for poultry [16, 17]. Group 1 consists of human coronaviruses, porcine transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), feline infectious peritonitis virus (FIPV) and some bat coronaviruses. Group 2 includes human coronavirus, bovine coronavirus, murine hepatitis virus, porcine hemagglutinating encephalomyelitis virus and bat coronaviruses. Group 3 consists of infectious bronchitis virus and turkey coronavirus.
Recent studies from the People’s Republic of China have identified bats as the most likely source of all coronaviruses [27, 30, 36, 45, 48]. Bats represent approximately 20% of all living mammal species and are distributed worldwide [22]. Bats have wide dietary diversity, including small vertebrates, nectar, pollen, fruits, blood, and insects. The majority of bat species live in social groups ranging from a few up to 20 million, the largest contiguous colony of mammals on earth [11]. All bat species have nocturnal activity and characteristic roosting behavior during the day. In Slovenia, we can find up to 30 different bat species that are characteristic of northern countries as well as species that are characteristic of southern countries, such as Rhinolophus euryale [37]. Insects are the main food of bat species living in Slovenia. Bats occupy diverse ecologic niches in a large range of habitats. Roost places include cavities in tree trunks, caves, cervices in rock and tree bark, bridges, cellars, wells and houses. During this time, they can have accidental contacts with humans, possibly enabling virus transmission.
In this study, we investigated whether bats in Slovenia also harbor coronaviruses. To our knowledge, this is the first report of coronaviruses in bats in Slovenia. One hundred six fecal samples from bats in Slovenia were analyzed using primers for the highly conserved RNA polymerase gene. Sequence comparisons were done with available sequences in GenBank database.
Materials and methods
Sample collection
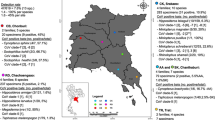
During the period from May to October 2008, a total 106 bats of 7 different species were captured at 27 different locations in Slovenia (Fig. 1). Bats were captured manually by using nets and hand nets. Live bats were captured in 16 different churches, 1 house, 1 castle, and 1 cave and by net trap at 8 different streams. All of the captured animals were placed separately into cotton bags for several minutes to calm them down before further investigations and sampling started. While being kept in the bag, the bats produced fecal pellets that were collected and transported from the field to the laboratory and stored at −70°C until use. Bats were classified according to species, sex, age, gravidity, and lactation status and typed by morphologic criteria by bat biologists. All manipulations of live animals were done in a manner consistent with our national guidelines for the capture, handling and care of bats.
Locations of sampling shown on a map of Slovenia. Each number (1–27) represents one location where bat samples were collected. Coronavirus RT-PCR-positive samples were found at the following locations: Spodnji Log in commune Kočevje (14), Livold in commune Kočevje (15), Tinje in commune Slovenska Bistrica (17), Knežja Njiva in commune Loška dolina (22) and Mali Otok in commune Postojna (23). Other sampling locations are Vnanje Gorice (1), Verd (2), Goče (3), Krašnja (4), Volovščica (5), Trebnje (6), Dolnji Suhor (7), Kobiljača (8), Završe (9), Kobilje (10), Mali Obrh (11), Močnik (12), Račna (13), Škocjan (16), Puščava (18), Dobrova (19), Selca (20), Snežnik (21), Velike Lašče (24), Iška vas (25), Cerkno (26) and Rižana (27)
Processing and analysis of samples
Fecal samples were suspended in minimum medium (RPMI 1640, Gibco) and homogenized by vortexing. Centrifugation was done at 3000 rpm for 10 min to obtain supernatants. From these supernatants, 50 μl of each sample was transferred to 500 μl of buffer AVL from the QIAamp Viral RNA Mini Kit (Qiagen) and processed further according to the instructions of the manufacturer. Total RNA was eluted in 50 μl of AVE buffer and was used as the template for reverse transcription and polymerase chain reaction (RT-PCR).
Electron microscopy
A limited number of samples (n = 16) that were in sufficient quantity was also examined for viruses using negatively stained direct electron microscopy (EM) of prepared stool suspension. The EM grid was incubated with 50 μl of stool suspension without a prior concentration step, washed with sterile water and incubated with 2% phosphotungstic acid (pH 4.5) for negative staining. Samples were examined using a JEM-1200 EX II transmission electron microscope (JEOL, Japan) for 20 min per grid at various sites prior to final result.
RT-PCR and sequencing
RT-PCR was performed by amplifying a 440-bp-long fragment of the highly conserved RNA-dependent RNA polymerase (pol) gene using the primers 5′ GGT TGG GAC TAT CCT AAG TGT GA 3′ and 5′ CCA TCA TCA GAT AGA ATC ATC ATA 3′, which were designed to amplify all known coronaviruses [43]. The RT-PCR was performed by using One Step RT-PCR Kit (Qiagen). The amplified products were analyzed by 1.8% agarose in gel electrophoresis and visualized under UV light after ethidium bromide staining. RT-PCR products from 14 positive samples were gel-purified and subjected to nucleotide sequence analysis. Two PCR products were sequenced for each strain. Sequencing reactions were performed using an ABI 310 Genetic Analyzer (Applied Biosystems) with ABI PRISM dye terminator sequencing chemistry, according to the manufacturer’s instructions.
Data analysis
Nucleotide sequences obtained from the RT-PCR products were assembled with the DNASTAR program and compared to the known sequences of the pol genes of coronaviruses in the GenBank database, using the Basic Local Alignment Search Tool (BLAST) program. A 405-bp region of the sequences obtained, which was present in most of the GenBank entries, was used for phylogenetic analysis. A multiple alignment of the nucleotide and protein coding sequences was constructed using Clustal W to determine genetic relationships among all of the sequences using neighbor-joining criteria. Final phylogenetic tree construction was performed with the Phylip program [12].
Results
From May to October 2008, live bats were caught and classified according to species, age, gravidity and lactation status (Table 1). A total of 106 fecal samples from 7 bat species were collected. To identify coronaviruses from these samples, an RT-PCR method using a primer pair from a conserved region was used to screen the field samples. Positive PCR products were detected in 14 out of 106 fecal samples examined. Coronavirus-positive bats were found at 5 different locations in Slovenia (Fig. 1) with 100% virus prevalence in Spodnji Log, 66.6% virus prevalence in Livold and Tinje, 33.3% virus prevalence in Mali Otok and 20% virus prevalence in Knežja njiva. Of the seven bat species studied, only one species, horseshoe bat (Rhinolophus hipposideros), was positive for coronaviruses (Table 1). Fourteen (38.8%) out of 36 horseshoe bat fecal samples were positive by RT-PCR. Four of the positive samples were from young males and ten of them were collected from females. Six of the positive females were classified as lactating, and four of the positive females were classified as young females.
The amplicon of each positive sample was subjected to DNA sequencing. To characterize the overall diversity of coronavirus sequences, including 14 sequences from this study (Table 2), a phylogenetic tree (Fig. 2) of a 405-bp fragment of the pol gene was generated from available sequences from the National Center for Biotechnology Information database (NCBI). Analysis of nucleotide and amino acid sequences of this region revealed that Slovenian coronavirus strains from this study belong to the group 2 coronaviruses. The grouping of other coronaviruses into three main groups (Group 1, Group 2, and Group 3) was done according to the previously defined groups. When comparing the 14 sequences obtained, the Slovenian strains were found to have 100% amino acid identity and 99.5–100% nucleotide identity. The obtained sequences of 12 strains form group 2 (Fig. 2) and share 60.8–85% nucleotide identity to our sequences. When compared to the closest previously characterized coronavirus in GenBank, the Slovenian strains share 85% nucleotide identity and 95.6% amino acid identity to strain Rp3/2004 (DQ071615). The genetic group of 14 coronaviruses from Slovenia forms a separate cluster with a significant bootstrap value (100%) within Group 2 (Fig. 2).
Phylogenetic tree based on nucleotide sequences of a partial pol gene sequence with representative isolates from bats, other mammals and birds (Table 2). Analyses were conducted using the PHYLIP program with partial sequences (405 nt) of each isolate taken from GenBank (corresponding to nt 15,234–15,638 of the genome of bat strain Rp3/2004 (DQ071615), using the neighbor-joining algorithm and a bootstrap test of phylogeny. Boostrap values were obtained from 1,000 datasets, and only nodes with bootstrap values higher than 70% are presented. The new group of 14 coronaviruses from this study was found to belong to Group 2 (bootstrap value 100%)
As described in “Materials and methods”, 16 samples were examined by EM, of which 10 where previously characterized as RT-PCR positive for coronaviruses. There was a limited amount of stool suspension, and thus it was not possible to perform any concentration technique prior to the EM examination. A clear coronavirus structure was detected in four of the coronavirus RT-PCR-positive samples examined (Fig. 3).
Discussion
To our knowledge, this is the first report of coronaviruses in bats in Slovenia. With more than 1,100 species, bats are among the most divergent and widely distributed mammals [41]. Bats are reservoirs for rabies virus and other lyssaviruses and were recently shown to be reservoirs for other important emerging viruses. More than 60 different RNA viruses have already been detected in bats, thus suggesting an important role in transmitting and maintaining zoonotic viruses [4, 20, 51]. It is known that Old World fruit bats are a reservoir for Hendra virus, which can cause severe respiratory illness in humans and horses [13, 23, 25, 32, 33, 35, 38, 40] and Nipah virus, which can cause lethal encephalitis and respiratory illness in humans and pigs [6, 7, 10, 18]. Bats can also be reservoirs for Ebola virus and Marburg viruses [2, 28].
The need for understanding the transmission, ecology and evolution of coronaviruses in wildlife was highlighted by a study showing that SARS coronaviruses caused large human epidemics in 2003. Those isolates were more closely related to isolates from palm civets than to the human epidemic strain of SARS coronavirus [42]. Sequence analyses of SARS coronavirus genomes after the epidemic suggested that multiple independent species-jumping events of SARS coronavirus from wild animals to humans have occurred [29, 42]. Interspecies transmissions of animal viruses to humans are a constant threat to human health. In order to obtain a better understanding of the prevalence of coronaviruses in our country, we collected 106 fecal samples from 7 different bat species.
To understand how coronaviruses might be transmitted and maintained in bat population, factors such as species, age, sex, gravidity and lactation status were observed. Out of 14 coronavirus-positive horseshoe bats, 4 of them were young males, 6 were lactating females, and 4 were young females. These results confirm that young bats of both sexes can be virus positive, as well as lactating bats, but virus was not detected in gravid bats. The data obtained in our study are similar to those from studies conducted in Germany [15]. Virus can be transmitted between young bats and lactating mothers, and from these data, it is highly possible that young bats provide a susceptible population for amplifying the coronavirus and transmitting it to lactating females in maternity colonies. Comparable to the experience with respiratory infections in humans, virus would replicate less efficiently in adults than in young animals because of partial immune protection, which is the result of infection in earlier life. This could explain the lower prevalence of virus in adult bats.
From the results of this study, we can speculate that our data do not reveal a complete picture of the presence of coronaviruses in bats in Slovenia because the RT-PCR method relies on detection of viral sequences. Animals that have already recovered from coronavirus infection could be negative in our assay but positive for antibodies. While sample sizes of each animal species are limited, viruses that are circulating at a low copy number may be missed. However, detecting coronaviruses by direct electron microscopy without prior concentration shows that a high concentration of coronaviruses could be excreted from bats’ feces. According to the estimated minimal particle concentration for EM diagnostics, it can be presumed that the coronavirus concentration in EM-positive samples is above 106 particles/ml [21]. The conserved primers used in our study were based on available coronavirus sequences and were used successfully for amplification of this new group of coronaviruses. However, these primers might not be able to detect coronaviruses that are genetically more divergent from previously known coronaviruses.
The new group of bat coronavirus sequences had typical features of coronaviruses. All Slovenian sequences had the highest percentage of amino acid identity with the group 2 coronaviruses. When compared with other bat isolates from group 2 coronaviruses (Rf1/2004, Rp3/2004, HKU3, Rm1/2004, HKU9-1), the Slovenian strains had from 75.6 to 96.3% amino acid identity and from 64% up to 85.8% nucleotide identity. The novel genetic group of coronaviruses presented in our study is probably the result of the first detection of coronaviruses in bats on our territory, and the high diversity of these viruses, resulting from the high mutation rate of RNA viruses due to infidelity of their polymerases and a higher chance of recombination as a result of their unique replication mechanism [55]. Before the SARS epidemic in 2003, a total of 19 coronaviruses were known, 2 of which were isolated from humans, 13 from other mammals and 4 from birds. Since the epidemic, 2 novel human [14] and at least 10 previously unrecognized coronaviruses from bats have been described [53]. The diversity of coronaviruses in bats is astonishing, and there are probably many other unknown coronaviruses in other animal species.
A recent phylogenetic analysis of coronaviruses in bats, other mammals, humans and birds has suggested that bats could be original host from which all coronavirus lineages are derived [54]. The first detection of coronaviruses from horseshoe bats in Slovenia and genetic analysis of sequences obtained from them has confirmed that viruses from this study are related to SARS coronaviruses from group 2 isolated from human. Horseshoe bats commonly roost in churches, houses and castles, where humans can be exposed and ecologic overlap between bats and humans can occur.
In conclusion, it is possible that another epidemic caused by an emerging coronavirus originating from bats could occur in the future. Detection, isolation and molecular epidemiology studies of bats in various geographic locations and animal species could help to elucidate host range, receptor specificities and the potential of coronaviruses in new emerging infectious diseases.
References
Almazan F, Gonzalez JM, Penzes Z, Izeta A, Calvo E, Plana-Duran J, Enjuanes L (2000) Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA 97(10):5516–5521
Biek R, Walsh PD, Leroy EM, Real LA (2006) Recent common ancestry of Ebola Zaire virus found in a bat reservoir. PLoS Pathog 2:90
Brierley I, Boursnell ME, Binns MM, Bilimoria B, Blok VC, Brown TD, Inglis SC (1987) An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J 6(12):3779–3785
Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T (2006) Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev 19:531–545
Cavanagh D (1997) Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol 142:629–633
Centers for Disease Control and Prevention. Outbreak of Hendra-like virus-Malaysia and Singapore (1998–1999) MMWR Morb Mortal Wkly Rep 48:265–269
Centers for Disease Control and Prevention (1999) Update: outbreak of Nipah virus-Malaysia and Singapore. MMWR Morb Mortal Wkly Rep 48:335–337
Chouljenko VN, Lin XQ, Storz J, Kousoulas KG, Gorbalenya AE (2001) Comparison of genomic and predicted amino acid sequences of respiratory and enteric bovine coronaviruses isolated from the same animal with fatal shipping pneumonia. J Gen Virol 82(PT12):2927–2933
Chu DK, Peiris JS, Chen H, Guan Y, Poon LL (2008) Genomic characterizations of bat coronaviruses (1A, 1B and HKU8) and evidence for co-infections in Miniopterus bats. J Gen Virol 89:1282–1287
Chua KB, Goh KJ, Wong KT, Kamarulzaman A, Tan PS, Ksiazek TG, Zaki RS, Paul G, Lam SK, Tan CT (1999) Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 354:1257–1259
Davis RB, Herreid CF, Short HL (1962) Mexican free tailed bats in Texas. Ecol Monogr 32:311–346
Felsenstein J (1989) Phylip phylogeny inference package (version 3.2). Cladistics 5:164–166
Field HE, Barratt PC, Hughes RJ, Shield J, Sullivan ND (2000) A fatal case of Hendra virus infection in horse in north Queensland: clinical and epidemiological features. Aust Vet J 78:279–280
Fouchier RA, Hartwig NG, Bestbroer TM, Nimeyer B, de Jong JC, Simon JH, Osterhaus AD (2004) A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci USA 101:6212–6216
Gloza-Rausch F, Ipsen A, Seebens A, Göttsche M, Panning M, Drexler JF, Petresen N, Annan A, Grywna K, Müller M, Pfefferle S, Drosten C (2008) Detection and prevalence patterns of Group I Coronaviruses in Bats, Northern Germany. Emerg Infect Dis 14:626–631
Gonzalez JM, Gomez-Puertas P, Cavanagh D, Gorbalenya AE, Enjuanes L (2003) A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch Virol 148:2207–2235
Gorbalenya AE, Snijder EJ, Spaan WJ (2004) Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J Virol 78:7863–7866
Goud AR (1996) Comparison of the deduced matrix and fusion protein sequences of equine morbillivirus with cognate genes of the Paramyxoviridae. Virus Res 43:17–31
Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Lui SW, Li PH, Zhang LJ, Guan Yj, Butt KL, Wong KL, Chan KW, Lim W, Shortridge KF, Yuen KY, Peiris JS, Poon LL (2003) Isolation and chracterisation of viruses related to SARS coronavirus from animals in shouter China. Science 302:276–278
Halpin K, Hyatt AD, Plowright RK, Epstein JH, Daszak P, Field HE, Wang L, Daniels PW, Henipavirus Ecology Research Group (2007) Emerging viruses: coming in on a wrinkled wing and a prayer. Clin Infect Dis 44:711–717
Hazelton PR, Gelderblom HR (2003) Electron microscopy for rapid diagnosis of infectious agents in emerging situations. Emerg Infect Dis 9(3):293–303
Holmes KV, Enjuanes L (2003) The SARS coronavirus: a postegomic era. Science 300:4508–4514
Hooper PT, Goud AR, Rusell GM, Kattenbelt JA, Mitchel G (1996) The retrospective diagnosis of a second outbreak of equine morbilivirus infection. Aust Vet J 74:244–245
Kocherhans R, Bridgen A, Tobler K (2001) Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes 23(2):137–144
Lai MM, Baric RS, Brayton PR, Stohlman SA (1984) Characterisation of leader RNA sequences on the virion and mRNA’s of muose hepatitis virus, a cytoplasmic RNA virus. Proc Natl Acad Sci USA 81:3626–3630
Lai MM, Cavanagh D (1997) The molecular biology of coronaviruses. Adv Virus Res 48:1–100
Lau SKP, Woo PCY, Li SKM, Huang Y, Tsoi HW, Wong BHL, Wong SSY, Leung SY, Chan KH, Yuen KY (2005) Severe acute respiratory syndrome coronavirus like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA 102:14040–14045
Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Delicat A, Paweska JT, Gonzalez JP, Swanpoel R (2005) Fruit bats as reservoirs of Ebola virus. Nature 438:575–576
Li W, Zhang C, Sui J, Kuhn JH, Moore MJ, Luo S, Wong SK, Huang IC, Xu K, Vasilieva N, Murakami A, He Y, Marasco WA, Guan Y, Choe H, Farzan M (2005) Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J 24:17634–17643
Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEchern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF (2005) Bats are natural reservoirs of SARS-like corinaviruses. Science 310:676–679
Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, Khattra J, Asano JK, Barber SA, Chan SY, Cloutier A, Coughlin SM, Freeman D, Girn N, Griffith OL, Leach SR, Mayo M, McDonald H, Montgomery SB, Pandoh PK, Petrescu DE, Robertson AG, Schein JE, Siddiqui A, Smailus DE, Stott DE, Yang GS, Plummer F, Andonov A, Artsob H, Bastien N, Bernard K, Booth TF, Bowness D, Czub M, Drebot M, Fernando L, Flick R, Garbutt M, Gray M, Grolla A, Jones S, Feldmann H, Meyers A, Kabani A, Li Y, Normand S, Stroher U, Tipples GA, Tyler S, Vogrig R, Ward D, Watson B, Brunham RC, Krajden M, Petric M, Skowronski DM, Upton C, Roper RL (2003) The genome sequence of the SARS-associated coronavirus. Science 300(5624):1399–1404
Murray K, Selleck P, Hooper P, Hyatt A, Gould A, Gleeson L, Westerbury H, Hiley L, Selvey L, Roodwell B, Ketterer P (1995) A morbilivirus that caused fatal disease in horses and humans. Science 268:94–97
Murray K, Rogers R, Salvey L, Selleck P, Hyatt A, Gould A, Gleeson L, Hooper P, Wesbury H (1995) A novel morbilivirus pneumonia of horses and its transmission to humans. Emerg Infect Dis 1:31–33
Novas-Martin S, Weiss SR (2003) SARS: lesions learned from other coronaviruses. Viral Immunol 16:461–474
O’Sullivan JD, Am Allworth, Paterson DL, Snow TM, Boots R, Gleeson LJ, Goud AR, Hyatt AD, Bradfield J (1997) Fatal encephalitis due to novel paramyxovirus transmitted from horses. Lancet 349:93–95
Poon LL, Chu DK, Chan KH, Wong OK, Ellis TM, Leung Yh, Lau SKP, Woo PCY, Suen KY, Yuen KY, Guan Y, Peiris JSM (2005) Identification of novel coronavirus in bats. J Virol 79:2001–2009
Presetnik P, Podgorelec M (2008) Ugotavljanje prisotnosti lyssavirusov pri netopirjih v letu 2008. Center za kartografijo favne in flore, Miklavž na Dravskem polju, p 1–27
Rogers RJ, Douglas IC, Badock FC, Glanville RJ, Seppanen KT, Gleeson LJ, Selleck PN, Dunn KJS (1996) Investigation of a second focus of equine morbillivirus infection in coastal Queensland. Aust Vet J 74:243–244
Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen MH, Tong S, Tamin A, Lowe M, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TC, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, NcCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus AD, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ (2003) Characterization of novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394–1399
Selvey LA, Wells RM, McCormack JG, Ansford AJ, Murray K, Rogers RJ, Lavercombe PS, Selleck P, Sheridan JW (1995) Infection of humans and horses by a newly described morbillivirus. Med J Aust 162:642–645
Simmons NB (2005) Order Chiroptera. In: Wilson DE, Reeder DM (eds) Mammal species of the world: a taxonomic and geographic reference. Johns Hopkins University Press, Baltimore, pp 312–529
Song HD, Tu CC, Zhang GW, Wang SY, Zheng K, Lei LC, Chen QX, Gao YW, Zhou HQ, Xiang H, Zheng HJ, Chern SW, Cheng F, Pan CM, Xuan H, Chen SJ, Luo HM, Zhou DH, Liu YF, He JF, Qin PZ, Li LH, Ren YQ, Liang WJ, Yu YD, Anderson L, Wang M, Xu RH, Wu XW, Zheng HY, Chen JD, Liang G, Gao Y, Liao M, Fang L, Jiang LY, Li H, Chen F, Di B, He LJ, Lin JY, Tong S, Kong X, Du L, Hao P, Tang H, Bernini A, Yu XJ, Spiga O, Guo ZM, Pan HY, He WZ, Manuguerra JC, Fontanet A, Danchin A, Niccolai N, Li YX, Wu CI, Zhao GP (2005) Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci USA 102:2430–2435
Stephensen CB, Casebolt DB, Gangpadhyay NN (1999) Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronaviruses and development of a consensus polymerase chain reaction assay. Virus Res 60(2):181–189
Sun L, Zhang GH, Jiang JW, Fu JD, Ren T, Cao WS, Xin CA, Liao M, Liu WJ (2007) A Massachusetts prototype like coronavirus isolated from wild peafowls is pathogenic to chickens. Virus Res 130(1–2):121–128
Tang XC, Zahng JX, Zhang YS, Wang P, Fan XH, Li LF, Li G, Dong BQ, Liu W, XU KM, Cheung CL, Song WJ, Vijaykrishna D, Pool LL, Peiris JS, Smith GJ, Chen H, Guan Y (2006) Prevalence of genetic diversity of coronaviruses in bats from China. J Virol 80:7481–7490
Thiel V, Herold J, Schelle B, Siddell SG (2001) Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J Gen Virol 82(PT6):1273–1281
van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wetheim-van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B (2004) Identification of a new human coronavirus. Nat Med 10:368–373
Vijaykrishna D, Smith GJ, Zhang JX, Peiris JS, Chen H, Guan Y (2007) Evolutionary insights into the ecology of coronaviruses. J Virol 81:4012–4020
Vijgen L, kayaerts E, Moes E, Thoelen I, Wollants E, Lemey P, Vandamme A, Van Renst M (2005) Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol 79(3):1595–1604
Vijgen L, Kayaerta E, Lemey P, Maes P, Van Reeth K, Nauwynck H, Pensaert M, Van Renst M (2006) Porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus. J Virol 80(14):7270–7274
Wong S, Lau S, Woo P, Yuen KY (2007) Bats as a continuing source of emerging infections in humans. Rev Med Virol 17:67–91
Woo PC, Lau SKP, Chan KH, Tsoi HW, Huang Y, Wong BHL, Cai JJ, Wong SSY, Peiris JSM, Chuc CM, Yuen KY (2005) Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol 79(2):884–895
Woo PC, Lau SK, Li KS, Poon RW, Wong BH, Tsoi HW, Yip BC, Huang Y, Chan KH, Yuen KY (2006) Molecular diversity of coronaviruses in bats. Virology 351:180–187
Woo PC, Wang M, Lau SK, Xu H, Poon RW, Guo R (2007) Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J Virol 81:1574–1585
Wu HY, Guy JS, Yoo D, Vlasak R, Urbach E, Brian DA (2003) Common RNA replication signals exist among group 2 coronaviruses: evidence for in vivo recombination between animal and human coronavirus molecules. Virology 315:174–183
Acknowledgments
We thank P. Presetnik, M. Podgorelec and M. Cipot for field sampling.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rihtarič, D., Hostnik, P., Steyer, A. et al. Identification of SARS-like coronaviruses in horseshoe bats (Rhinolophus hipposideros) in Slovenia. Arch Virol 155, 507–514 (2010). https://doi.org/10.1007/s00705-010-0612-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-010-0612-5