Abstract
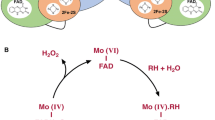
The first crystal structure of mammalian monoamine oxidases (MAOs) was solved in 2002; almost 65 years after, these FAD-dependent enzymes were discovered and classified as responsible for the oxidation of aromatic neurotransmitters. Both MAO A and MAO B feature a two-domain topology characterized by the Rossmann fold, interacting with dinucleotide cofactors, which is intimately associated to a substrate-binding domain. This globular body is endowed with a C-terminal α-helix that anchors the protein to the outer mitochondrial phospholipid bilayer. As monotopic membrane proteins, the structural elucidation of MAOs was a challenging task that required the screening of different detergent conditions for their purification and crystallization. MAO A and MAO B structures differ both in their oligomerization architecture and in details of their active sites. Purified human MAO B and rat MAO A are dimeric, whereas human MAO A was found to be monomeric, which is believed to result from the detergent treatments used to extract the protein from the membrane. The active site of MAOs consists of a hydrophobic cavity located in front of the flavin cofactor and extending to the protein surface. Some structural features are highly conserved in the two isozymes, such as a Tyr–Tyr aromatic sandwich in front of the flavin ring and a Lys residue hydrogen-bonded to the cofactor N5 atom, whereas a pair of gating residues (Phe208/Ile335 in MAO A; Ile199/Tyr326 in MAO B) specifically determines the different substrate and inhibitor properties of the two enzymes.




(source: Membrane Proteins of Known 3D Structures, http://blanco.biomol.uci.edu/mpstruc/) is depicted within its native membrane compartment, either in the prokaryotic (left) or in the eukaryotic (right) cell



Similar content being viewed by others
Abbreviations
- MAO:
-
Monoamine oxidase
- FAD:
-
Flavin adenine dinucleotide
- ROS:
-
Reactive oxygen species
References
Bailey SD, Bucci L, Gosline E, Kline NS, Park IH, Rochlin D, Saunders JC, Vaisberg M (1959) Comparison of iproniazid with other amine oxidase inhibitors, including W-1544, JB-516, RO 4-1018, and RO 5-0700. Ann N Y Acad Sci 80:652–668
Binda C, Newton-Vinson P, Hubalek F, Edmondson DE, Mattevi A (2002) Structure of human monoamine oxidase B, a drug target for the treatment of neurological disorders. Nat Struct Biol 9(1):22–26. https://doi.org/10.1038/nsb732
Binda C, Wang J, Pisani L, Caccia C, Carotti A, Salvati P, Edmondson DE, Mattevi A (2007) Structures of human monoamine oxidase B complexes with selective noncovalent inhibitors: safinamide and coumarin analogs. J Med Chem 50(23):5848–5852. https://doi.org/10.1021/jm070677y
Binda C, Valente S, Romanenghi M, Pilotto S, Cirilli R, Karytinos A, Ciossani G, Botrugno OA, Forneris F, Tardugno M, Edmondson DE, Minucci S, Mattevi A, Mai A (2010) Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2. J Am Chem Soc 132(19):6827–6833. https://doi.org/10.1021/ja101557k
Binda C, Milczek EM, Bonivento D, Wang J, Mattevi A, Edmondson DE (2011) Lights and shadows on monoamine oxidase inhibition in neuroprotective pharmacological therapies. Curr Top Med Chem 11(22):2788–2796
Bonivento D, Milczek EM, McDonald GR, Binda C, Holt A, Edmondson DE, Mattevi A (2010) Potentiation of ligand binding through cooperative effects in monoamine oxidase B. J Biol Chem 285(47):36849–36856. https://doi.org/10.1074/jbc.M110.169482
Byrne B (2015) Pichia pastoris as an expression host for membrane protein structural biology. Curr Opin Struct Biol 32:9–17. https://doi.org/10.1016/j.sbi.2015.01.005
Caffrey M (2015) A comprehensive review of the lipid cubic phase or in meso method for crystallizing membrane and soluble proteins and complexes. Acta Crystallogr F Struct Biol Commun 71(Pt 1):3–18. https://doi.org/10.1107/S2053230X14026843
Cesura AM, Pletscher A (1992) The new generation of monoamine oxidase inhibitors. Prog Drug Res 38:171–297
Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC (2007) High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318(5854):1258–1265. https://doi.org/10.1126/science.1150577
de Kroon AI, Dolis D, Mayer A, Lill R, de Kruijff B (1997) Phospholipid composition of highly purified mitochondrial outer membranes of rat liver and Neurospora crassa. Is cardiolipin present in the mitochondrial outer membrane? Biochim Biophys Acta 1325(1):108–116
De Colibus L, Li M, Binda C, Lustig A, Edmondson DE, Mattevi A (2005) Three-dimensional structure of human monoamine oxidase A (MAO A): relation to the structures of rat MAO A and human MAO B. Proc Natl Acad Sci USA 102(36):12684–12689. https://doi.org/10.1073/pnas.0505975102
Deisenhofer J, Michel H (1989) Nobel lecture. The photosynthetic reaction centre from the purple bacterium Rhodopseudomonas viridis. EMBO J 8(8):2149–2170
Edmondson DE, Binda C (2018) Monoamine Oxidases. Subcell Biochem 87:117–139. https://doi.org/10.1007/978-981-10-7757-9_5
Edmondson DE, Binda C, Wang J, Upadhyay AK, Mattevi A (2009) Molecular and mechanistic properties of the membrane-bound mitochondrial monoamine oxidases. Biochemistry 48(20):4220–4230. https://doi.org/10.1021/bi900413g
Forneris F, Battaglioli E, Mattevi A, Binda C (2009) New roles of flavoproteins in molecular cell biology: histone demethylase LSD1 and chromatin. FEBS J 276(16):4304–4312. https://doi.org/10.1111/j.1742-4658.2009.07142.x
Fowler PW, Balali-Mood K, Deol S, Coveney PV, Sansom MS (2007) Monotopic enzymes and lipid bilayers: a comparative study. Biochemistry 46(11):3108–3115. https://doi.org/10.1021/bi602455n
Hunte C, Michel H (2002) Crystallisation of membrane proteins mediated by antibody fragments. Curr Opin Struct Biol 12(4):503–508
Kuhlbrandt W (2014) Cryo-EM enters a new era. Elife 3:e03678. https://doi.org/10.7554/eLife.03678
Lees AJ, Shaw KM, Kohout LJ, Stern GM, Elsworth JD, Sandler M, Youdim MB (1977) Deprenyl in Parkinson’s disease. Lancet 2(8042):791–795
Li M, Hubalek F, Newton-Vinson P, Edmondson DE (2002) High-level expression of human liver monoamine oxidase A in Pichia pastoris: comparison with the enzyme expressed in Saccharomyces cerevisiae. Protein Expr Purif 24(1):152–162. https://doi.org/10.1006/prep.2001.1546
Lin YC, Chang YT, Campbell M, Lin TP, Pan CC, Lee HC, Shih JC, Chang PC (2017) MAOA-a novel decision maker of apoptosis and autophagy in hormone refractory neuroendocrine prostate cancer cells. Sci Rep 7:46338. https://doi.org/10.1038/srep46338
Ma J, Yoshimura M, Yamashita E, Nakagawa A, Ito A, Tsukihara T (2004) Structure of rat monoamine oxidase A and its specific recognitions for substrates and inhibitors. J Mol Biol 338(1):103–114. https://doi.org/10.1016/j.jmb.2004.02.032
Maggiorani D, Manzella N, Edmondson DE, Mattevi A, Parini A, Binda C, Mialet-Perez J (2017) Monoamine oxidases, oxidative stress, and altered mitochondrial dynamics in cardiac ageing. Oxid Med Cell Longev 2017:3017947. https://doi.org/10.1155/2017/3017947
Magnani F, Serrano-Vega MJ, Shibata Y, Abdul-Hussein S, Lebon G, Miller-Gallacher J, Singhal A, Strege A, Thomas JA, Tate CG (2016) A mutagenesis and screening strategy to generate optimally thermostabilized membrane proteins for structural studies. Nat Protoc 11(8):1554–1571. https://doi.org/10.1038/nprot.2016.088
Manglik A, Kobilka BK, Steyaert J (2017) Nanobodies to study G protein-coupled receptor structure and function. Ann Rev Pharmacol Toxicol 57:19–37. https://doi.org/10.1146/annurev-pharmtox-010716-104710
McNicholas S, Potterton E, Wilson KS, Noble ME (2011) Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr 67(Pt 4):386–394. https://doi.org/10.1107/S0907444911007281
Mialet-Perez J, Santin Y, Parini A (2018) Monoamine oxidase-A, serotonin and norepinephrine: synergistic players in cardiac physiology and pathology. J Neural Transm (Vienna). https://doi.org/10.1007/s00702-018-1908-y
Milczek EM, Binda C, Rovida S, Mattevi A, Edmondson DE (2011) The ‘gating’ residues Ile199 and Tyr326 in human monoamine oxidase B function in substrate and inhibitor recognition. FEBS J 278(24):4860–4869. https://doi.org/10.1111/j.1742-4658.2011.08386.x
Newton-Vinson P, Hubalek F, Edmondson DE (2000) High-level expression of human liver monoamine oxidase B in Pichia pastoris. Protein Expr Purif 20(2):334–345. https://doi.org/10.1006/prep.2000.1309
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612. https://doi.org/10.1002/jcc.20084
Ramsay RR, Albreht A (2018) Kinetics, mechanism, and inhibition of monoamine oxidase. J Neural Transm (Vienna). https://doi.org/10.1007/s00702-018-1861-9
Rebrin I, Geha RM, Chen K, Shih JC (2001) Effects of carboxyl-terminal truncations on the activity and solubility of human monoamine oxidase B. J Biol Chem 276(31):29499–29506. https://doi.org/10.1074/jbc.M100431200
Reis J, Manzella N, Cagide F, Mialet-Perez J, Uriarte E, Parini A, Borges F, Binda C (2018) Tight-binding inhibition of human monoamine oxidase B by chromone analogs: A kinetic, crystallographic, and biological analysis. J Med Chem 61(9):4203–4212. https://doi.org/10.1021/acs.jmedchem.8b00357
Riederer P, Muller T (2017) Use of monoamine oxidase inhibitors in chronic neurodegeneration. Expert Opin Drug Metab Toxicol 13(2):233–240. https://doi.org/10.1080/17425255.2017.1273901
Shih JC (1991) Molecular basis of human MAO A and B. Neuropsychopharmacology 4(1):1–7
Son SY, Ma J, Kondou Y, Yoshimura M, Yamashita E, Tsukihara T (2008) Structure of human monoamine oxidase A at 2.2—a resolution: the control of opening the entry for substrates/inhibitors. Proc Natl Acad Sci USA 105(15):5739–5744. https://doi.org/10.1073/pnas.0710626105
Tatton W, Chalmers-Redman R, Tatton N (2003) Neuroprotection by deprenyl and other propargylamines: glyceraldehyde-3-phosphate dehydrogenase rather than monoamine oxidase B. J Neural Transm (Vienna) 110(5):509–515. https://doi.org/10.1007/s00702-002-0827-z
Tipton KF (2018) 90 years of monoamine oxidase: some progress and some confusion. J Neural Transm (Vienna). https://doi.org/10.1007/s00702-018-1881-5
Upadhyay AK, Edmondson DE (2009) Development of spin-labeled pargyline analogues as specific inhibitors of human monoamine oxidases A and B. Biochemistry 48(18):3928–3935. https://doi.org/10.1021/bi9002106
Weyler W, Hsu YP, Breakefield XO (1990) Biochemistry and genetics of monoamine oxidase. Pharmacol Ther 47(3):391–417
Youdim MB, Kupershmidt L, Amit T, Weinreb O (2014) Promises of novel multi-target neuroprotective and neurorestorative drugs for Parkinson’s disease. Parkinsonism Relat Disord 20(Suppl 1):S132–S136. https://doi.org/10.1016/S1353-8020(13)70032-4
Zeller EA (1938) Über den enzymatischen Abbau von Histamin und Diaminen. 2. Mitteilung Helv Chim Acta 21 (1):880–890
Acknowledgements
This work was supported by Fondazione Cariplo (Grant no. 2014 − 0672 to C.B.) and by the Italian Ministry of Education, University and Research (MIUR, “Dipartimenti di Eccellenza Program 2018–2022—Dept. of Biology and Biotechnology L. Spallanzani”, University of Pavia).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Iacovino, L.G., Magnani, F. & Binda, C. The structure of monoamine oxidases: past, present, and future. J Neural Transm 125, 1567–1579 (2018). https://doi.org/10.1007/s00702-018-1915-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00702-018-1915-z