Abstract
Background
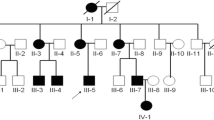
Arachnoid cysts are congenital fluid-filled compartments within the cerebrospinal fluid cisterns and cerebral fissures. They most commonly occur sporadically, and familial occurrence has rarely been reported. In this study, we showed the first genetic linkage in the literature in a pure intracranial arachnoid cyst family with autosomal recessive trait.
Methods
We identified an intracranial arachnoid cyst family in southern Turkey whose six of seven offspring had intracranial arachnoid cysts in different localizations, and collected venous blood from seven offspring of the family. Whole-genome linkage analysis was performed in all offspring.
Results
A theorical maximum logarithm of the odds score of 4.6 was identified at chromosome 6q22.31-23.2. This result shows strong genetic linkage to this locus.
Conclusions
We present the first genetic linkage analysis result in a pure intracranial arachnoid cyst family in literature. Further investigation of this linkage area can reveal a causative gene causing the intracranial arachnoid cyst phenotype and can illuminate the pathogenesis of this disease.




Similar content being viewed by others
References
Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101
Allen A, Wiegmann TB, MacDougall ML (1986) Arachnoid cyst in a patient with autosomal-dominant polycystic kidney disease. Am J Kidney Dis 8:128–130
Bayrakli F, Bilguvar K, Mason CE, DiLuna ML, Bayri Y, Gungor L, Terzi M, Mane SM, Lifton RP, State MW, Gunel M (2007) Rapid identification of disease-causing mutations using copy number analysis within linkage intervals. Hum Mutat 28:1236–1240
Bilguvar K, Ozturk AK, Bayrakli F, Guzel A, DiLuna ML, Bayri Y, Tatli M, Tekes S, Arlier Z, Yasuno K, Mason CE, Lifton RP, State MW, Gunel M (2009) The syndrome of pachygyria, mental retardation, and arachnoid cysts maps to 11p15. Am J Med Genet A 149A:2569–2572
Bittles AH, Black ML (2010) Evolution in health and medicine Sackler colloquium: Consanguinity, human evolution, and complex diseases. Proc Natl Acad Sci USA 107(Suppl 1):1779–1786
Guzel A, Tatli M, Bilguvar K, Diluna ML, Bakkaloglu B, Ozturk AK, Bayrakli F, Gunel M (2007) Apparently novel genetic syndrome of pachygyria, mental retardation, seizure, and arachnoid cysts. Am J Med Genet A 143:672–677
Handa J, Okamoto K, Sato M (1981) Arachnoid cyst of the middle cranial fossa: report of bilateral cysts in siblings. Surg Neurol 16:127–130
Jamjoom ZA, Okamoto E, Jamjoom AH, al-Hajery O, Abu-Melha A (1995) Bilateral arachnoid cysts of the Sylvian region in female siblings with glutaric aciduria type I. Report of two cases. J Neurosurg 82:1078–1081
Kanev P (2004) Arachnoid Cysts. In: Winn H (ed) Youmans Neurological Surgery. vol 3. Saunders, pp 3289–3299
Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247
Masuno M, Fukushima Y, Sugio Y, Kuroki Y (1987) Partial distal 12q trisomy with arachnoid cyst. Jinrui Idengaku Zasshi 32:39–43
McClintick J, Koller DL, Pankratz N, Kirkwood SC, Naughton B, Foroud T (2001) Parametric linkage analysis and disequilibrium methods to identify loci for complex disease. Genet Epidemiol 21(Suppl 1):S528–S533
Osborn AG, Preece MT (2006) Intracranial cysts: radiologic-pathologic correlation and imaging approach. Radiology 239:650–664
Palmer LJ, Schnell AH, Witte JS, Elston RC (2002) Parametric linkage analysis. Meth Mol Biol 195:13–35
Pomeranz S, Constantini S, Lubetzki-Korn I, Amir N (1991) Familial intracranial arachnoid cysts. Childs Nerv Syst 7:100–102
Rice JP, Saccone NL, Corbett J (2008) Model-based methods for linkage analysis. Adv Genet 60:155–173
Sinha S, Brown JI (2004) Familial posterior fossa arachnoid cyst. Childs Nerv Syst 20:100–103
Tadmouri GO, Nair P, Obeid T, Al Ali MT, Al Khaja N, Hamamy HA (2009) Consanguinity and reproductive health among Arabs. Reprod Health 6:17
Tolmie JL, Day R, Fredericks B, Galea P, Moffett AW (1997) Dominantly inherited cerebral dysplasia: arachnoid cyst associated with mild mental handicap in a mother and her son. J Med Genet 34:1018–1020
Conflicts of interest
None.
Author information
Authors and Affiliations
Corresponding author
Additional information
Comment
The authors present a consanguineous Turkish family in which both of the parents were unaffected but six of their seven children had an intracranial arachnoid cyst in MRI. Genome-wide linkage analysis (250K SNP array) suggested an interval 6q22.31-23.2 of nearly 13 Mb with a LOD score of 4.6. The area may or may not contain a causative rare mutation or variant exposed by the consanguinity. This small article is likely to pass busy neurosurgeons' eyes, but it touches several important topics in the genomics and epigenomics of the development and diseases of the human brain.
Most importantly, we think that arachnoid cysts are just bags of soft meninges filled with CSF, not predisposed genetically nor a complex trait - haha dead wrong - nothing is utterly random or chainsaw simple in the human tissues and organs nor in their diseases and malformations.
Human DNA contains some 20,000 genes, as young neurosurgeons learn in medical school. Do these genes contain enough data to blueprint all human tissues and organs that are hugely complex under macroscope and microscope? – no way near. How about alternative splicing producing hundreds of thousands of protein isoforms? – not better. In human brain tissue, each neuron may form hundreds of connections with fellow neurons, a computing network of unimaginable complexity. So where are the blueprints of all human macroscopic organ and microscopic tissue complexity that repeats from one generation to another? Genes make only 1–2% of the whole DNA sequence – so what is the rest of DNA doing? The rest has magnitudes of functions which are just starting to be uncovered.
And where are the undesired blueprints among the structural blueprints that predispose to complex traits such hypertension, TD2, Alzheimer's disease, or intracranial aneurysm, to the desire of risk factors such as smoking or alcohol, and to the tissue-specific susceptibilities and effects of the risk factors? – previously termed bluntly as 'environmental effects'. The variome of single nucleotide polymorphisms (SNPs) caused a huge hype, but rather little concrete was achieved from the colossal investment in gathering very large international cohorts of patients and controls and performing genome-wide SNP association studies (GWASs) in complex traits such as intracranial aneurysms (Yasuno 2011). Nevertheless, being one of a hundred or so authors in Nature Genetics (Impact Factor 36) articles increases the personal H-index and chances of research funding. So where is the hidden heritability of complex traits? Previous GWASs investigated common SNPs with minor allele frequency of >5%, so projects like One Thousand Genomes are cataloguing rarer variants, predictably for another round of the GWAS cohort mill.
One trend is to hunt even rarer causative variants that may become exposed in consanguineous pedigrees and isolates. Cousin marriages, often abhorred in the West, may make over half of all marriages in areas of the Middle East. This centuries-old tradition is not genetically detrimental, although it increases the incidence of recessive traits. Unintentionally, consanguineous marriages open a research window to identify very rare causative variants behind complex traits (Bilguvar 2010).
Hidden blueprints of the complexity in human structure and disease likely reside in the epigenome (3-5). Epigenomic marks such as DNA methylation or histone tail modifications do no alter the DNA sequence. These modifications may pass from one generation to another. The epigenome of the cell is highly dynamic, governed by a complex interplay of genetic and environmental factors. DNA methylation patterns are specific to various tissues and developmental stages, and they also change over time. The full range of epigenetic modifications in a cell is just astronomical.
It is about cells and brain on Earth. Young neurosurgeons, study epigenomics of the CNS tissues.
Juha E Jääskeläinen
Kuopio, Finland
References
1. Yasuno K, Bilguvar K, Bijlenga P, Low SK, Krischek B, Auburger G, Simon M, Krex D, Arlier Z, Nayak N, Ruigrok YM, Niemelä M, Tajima A, von und zu Fraunberg M, Dóczi T, Wirjatijasa F, Hata A, Blasco J, Oszvald A, Kasuya H, Zilani G, Schoch B, Singh P, Stüer C, Risselada R, Beck J, Sola T, Ricciardi F, Aromaa A, Illig T, Schreiber S, van Duijn CM, van den Berg LH, Perret C, Proust C, Roder C, Ozturk AK, Gaál E, Berg D, Geisen C, Friedrich CM, Summers P, Frangi AF, State MW, Wichmann HE, Breteler MM, Wijmenga C, Mane S, Peltonen L, Elio V, Sturkenboom MC, Lawford P, Byrne J, Macho J, Sandalcioglu EI, Meyer B, Raabe A, Steinmetz H, Rüfenacht D, Jääskeläinen JE, Hernesniemi J, Rinkel GJ, Zembutsu H, Inoue I, Palotie A, Cambien F, Nakamura Y, Lifton RP, Günel M (2010) Genome-wide association study of intracranial aneurysm identifies three new risk loci. Nat Genet 42(5):420-5
2. Bilgüvar K, Oztürk AK, Louvi A, Kwan KY, Choi M, Tatli B, Yalnizoğlu D, Tüysüz B, Cağlayan AO, Gökben S, Kaymakçalan H, Barak T, Bakircioğlu M, Yasuno K, Ho W, Sanders S, Zhu Y, Yilmaz S, Dinçer A, Johnson MH, Bronen RA, Koçer N, Per H, Mane S, Pamir MN, Yalçinkaya C, Kumandaş S, Topçu M, Ozmen M, Sestan N, Lifton RP, State MW, Günel M (2010) Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 9;467(7312):207-10
3. Gibson G (2012) Rare and common variants: twenty arguments. Nat Rev Genet 18;13(2):135-45
4. Feil R, Fraga MF (2012) Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet 4;13(2):97-109
5. Stower H. (2012) Epigenetics: Dynamic DNA methylation. Nat Rev Genet 4;13(2):75
This work was supported by the Scientific Research Project Fund of Cumhuriyet University under project number T-455.
Rights and permissions
About this article
Cite this article
Bayrakli, F., Okten, A.I., Kartal, U. et al. Intracranial arachnoid cyst family with autosomal recessive trait mapped to chromosome 6q22.31-23.2. Acta Neurochir 154, 1287–1292 (2012). https://doi.org/10.1007/s00701-012-1312-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00701-012-1312-6