Abstract
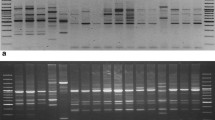
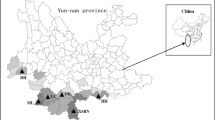
Genetic variation within and among population is the basis for survival of the population both in short and long term. Thus, studying the plant genetic diversity is essential for any conservation program. Indigenous medicinal plants like Justicia adhatoda L. which are facing high rate of depletion from the wild population need immediate attention. DNA-based dominant molecular marker techniques, random amplification of polymorphic DNA (RAPD) and inter-simple sequence repeat (ISSR) were used to unravel the genetic variability and relationships across thirty-two wild accessions of J. adhatoda L., a valuable medicinal shrub widespread throughout the tropical regions of Southeast Asia. Amplification of genomic DNA using 38 primers (18 RAPD and 20 ISSR) yielded 434 products, of which 404 products were polymorphic revealing 93.11 % polymorphism. The average polymorphic information content value obtained with RAPD and ISSR markers was 0.25 and 0.24, respectively. Marker index (RAPD = 3.94; ISSR = 3.53) and resolving power (RAPD = 4.24; ISSR = 3.94) indicate that the RAPD markers were relatively more efficient than the ISSR assay revealing the genetic diversity of J. adhatoda. The Shannon diversity index obtained with RAPD and ISSR markers was 0.40 and 0.38, respectively. The similarity coefficient ranged from 0.26 to 0.89, 0.33 to 0.93 and 0.31 to 0.90 with RAPD, ISSR and combined UPGMA dendrogram, respectively. PCA derived on the basis of pooled data of both the markers illustrated that the first three principal coordinate components accounted 79.27 % of the genetic similarity variance. The mantel test between two Jaccard’s similarity matrices gave r = 0.901, showing the fit correlation between ISSR- and RAPD-based similarities. Based on the results, ex-situ methods may be the most suitable and efficient measure for long-term conservation.








Similar content being viewed by others
References
Arif M, Zaidi NW, Singh YP, Haq QMR, Singh US (2009) A comparative analysis of ISSR and RAPD markers for study of genetic diversity in Shisham (Dalbergia sissoo). Plant Mol Biol Rep 27:488–495
Arnaud-Haond S, Teixeira S, Massa S, Billot C, Saenger P, Coupland G, Duarte C, Serrão E (2006) Genetic structure at range edge: low diversity and high inbreeding in Southeast Asian mangrove (Avicennia marina) populations. Mol Ecol 15:3515–3525
Atal CK (1980) Chemistry and pharmacology of vasicine: a new oxytocin and abortifacient. Indian Drugs 15:15–18
Byars SG, Parsons Y, Hoffmann AA (2009) Effect of altitude on the genetic structure of an Alpine grass, Poa hiemata. Ann Bot 103:885–899
Chakrabarty A, Brantner AH (2001) Study of alkaloids from Adhatoda vasica Nees on their anti-inflammatory activity. Phytother Res 15:532–534
Chen JM, Gituru WR, Wang YH, Wang QF (2006) The extent of clonality and genetic diversity in the rare Caldesia grandis (Alismataceae): comparative results for RAPD and ISSR markers. Aquat Bot 84:301–307
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size. Implications for plant conservation. Ann Rev Ecol Syst 24:217–242
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: applications to human mitochondrial DNA restriction data. Genetics 131:479–491
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Frankel OH, Brown AHD, Burdon JJ (1995) The conservation of plant biodiversity. Cambridge University Press, Cambridge
Gilani SA, Fujii Y, Kikuchi A, Shinwari ZK, Watanabe KN (2011) Ecological consequences, genetic and chemical variations in fragmented populations of a medicinal plant, Justicia adhatoda and implication for its conservation. Pak J Bot 43:29–37
Guo WL, Gong L, Ding ZF, Li YD, Li FX, Zhao SP, Liu B (2006) Genomic instability in phenotypically normal regenerants of medicinal plant Codonopsis lanceolata Benth. Et Hook. f., as revealed by ISSR and RAPD markers. Plant Cell Rep 25:896–906
Hamrick JL, Godt MJW (1989) Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant population genetics, breeding, and genetic resources 43–63. Sinauer, Sunderland, Massachusetts
Hamrick JL, Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Philos Trans R Soc Lond Ser B Biol Sci 351:1291–1298
Ince AG, Karaca M, Onus AN (2010) Genetic relationships within and between Capsicum species. Biochem Genet 48:83–95
Jaccard P (1908) Nouvelle recherches sur La distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Khanuja SPS, Shasany AK, Darokar MP, Kumar S (1999) Rapid isolation of DNA from dry and fresh samples of plants producing large amounts of secondary metabolites and essential oils. Plant Mol Bio Rep 17:1–7
Kiani M, Memariani F, Zarghami H (2012) Molecular analysis of species of Tulipa L. from Iran based on ISSR markers. Plant Syst Evol. doi:10.1007/s00606-012-0654-0
Li A, Ge S (2006) Genetic variation and conservation of Changnienia amoena, an endangered orchid endemic to China. Plant Syst Evol 258:251–260
Liu YF, Xing M, Zhao W, Fan RJ, Luo S, Chen X (2012) Genetic diversity analysis of Rhododendron aureum Georgi (Ericaceae) located on Changbai Mountain using ISSR and RAPD markers. Plant Syst Evol 298:921–930
Loveless MD, Hamrick JL (1984) Ecological determinants of genetic structure in plant populations. Ann Rev Ecol Syst 15:65–95
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Marshall DF (1997) Meeting training needs in developing countries. In: Ayad WG, Hodgkin T, Jaradat A, Rao VR (eds) Molecular genetic techniques for plant genetic resource. IPGRI, Rome, pp 128–132
McDermott JM, McDonald BA (1993) Gene flow in plant pathosystems. Annu Rev Phytopathol 31:353–373
Munoz M, Warner J, Albertazzi FJ (2010) Genetic diversity analysis of the endangered slipper orchid Phragmipedium longifolium in Costa Rica. Plant Syst Evol 290:217–223
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Nat Acad Sci 70:3321–3323
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York, pp 176–187
Ohsawa T, Ide Y (2008) Global patterns of genetic variation in plant species along vertical and horizontal gradients on mountains. Glob Ecol Biogeogr 17:152–163
Patel VK, Venkata-Krishna- Bhatt H (1984) In vitro study of antimicrobial activity of Adhatoda vasica (L.) (Leaf extract) on gingival inflammation-A preliminary report. Ind J Med Sci 38:70–72
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genetic 98:107–112
Qian W, Ge S, Hong DY (2001) Genetic variation within and among populations of a wild rice Oryza granulata from China detected by RAPD and ISSR markers. Theor Appl Genet 102:440–449
Raina SN, Rani V, Kojima T, Ogihara Y, Singh KP, Devarumath RM (2001) RAPD and ISSR fingerprints as useful genetic markers for analysis of genetic diversity, varietal identification and phylogenetic relationships in parent (Arachis hypogea) accessions and wild species. Genome 44:763–772
Rohlf FJ (1998) NTSYS-PC: numerical taxonomy and multivariate analysis system. Version 2.02, Exeter software. Setauket, New York
Roldan-Ruiz I, Dendauw J, Vanbockstaele E, Depicker A, De Loose M (2000) AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol Breed 6:125–134
Rossetto M, Weaver PK, Dixon KW (1995) Use of RAPD analysis in devising conservation strategies for the rare and endangered Grevillea scapigera (Proteaceae). Mol Eco 4:321–329
Shinwari ZK (2010) Medicinal plants research in Pakistan. J Med Plant Res 4(3):161–176
Shivanna KR (2009) Pollination biology, breeding system and reproductive success of Adhatoda vasica, an important medicinal plant. Curr Sci 96(3):408–412
Singh DR, Srivastava AK, Srivastava A, Srivastava RC (2011) Genetic diversity among three Morinda species using RAPD and ISSR markers. Ind J Biotech 10:285–293
Sundaram S, Purwar S (2011) Assessment of genetic diversity among fenugreek (Trigonella foenum-graecum L.), using RAPD molecular markers. J Med Plants Res 5(9):1543–1548
Tripathi N, Chouhan DS, Saini N, Tiwari S (2012) Assessment of genetic variations among highly endangered medicinal plant Bacopa monnieri (L.) from Central India using RAPD and ISSR analysis. 3. Biotech 2:327–336
Van De Peer Y, Wachter R (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comp Appl Biosci 10:569–570
Varshney RK, Chabane K, Hendre PS, Aggarwal RK, Graner A (2007) Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and elite barleys. Plant Sci 173:638–649
Verma S, Rana TS (2013) Genetic relationships among wild and cultivated accessions of curry leaf plant (Murraya koenigii (L.) Spreng.), as revealed by DNA fingerprinting methods. Mol Biotechnol 53:139–149
Williams LGK, Kubelik AR, Levak KJ et al (1990) DNA polymorphism amplification by arbitrary primers are useful as genetic markers. Nuc Acid Res 18:641–646
Yeh FC, Yang RC, Boyle TBJ, Ye ZH, Mao JX (1999) POPGENE 3.2, User-friendly shareware for population genetic analysis. http://ualberta.ca/wfeyeh
Zaghloul M, Reisch C, Poschlod P (2013) Soil seed bank contributes significantly to genetic variation of Hypericum sinaicum in a changing environment. Plant Syst Evol. doi:10.1007/s00606-013-0837-3
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183
Acknowledgments
The authors wish to express their sincere thanks to Director, CSIR-CIMAP (Lucknow), for providing the facilities to carry out this research. The financial support from the Council of Scientific and Industrial Research (CSIR), New Delhi, India through the projects (XIIth FYP) ChemBiO (BSC-0203) and BioprosPR (BSC-0106) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kumar, A., Mishra, P., Singh, S.C. et al. Efficiency of ISSR and RAPD markers in genetic divergence analysis and conservation management of Justicia adhatoda L., a medicinal plant. Plant Syst Evol 300, 1409–1420 (2014). https://doi.org/10.1007/s00606-013-0970-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-013-0970-z