Abstract.
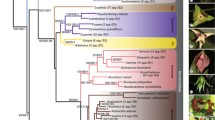
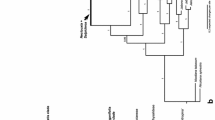
Cladistic analysis of rbcL nucleotide sequences was applied to 58 taxa representing most subgenera and sections of Luzula and Juncus, chosen to reflect morphological and geographical diversity of both genera. Additionally, representatives of all other genera of the Juncaceae and two taxa from the Cyperaceae were included. Phylogenetic trees were constructed using parsimony with Prionium serratum as outgroup. The dataset has 190 parsimony informative sites. The analysis yielded more than 332,400 equally parsimonious trees (length 620, CI=0.47, RI=0.82). A jackknife analysis revealed several well-supported clades. Luzula is monophyletic and Juncus is non-monophyletic. Each of the generally accepted subgenera of Juncus, subg. Juncus and subg. Agathryon, form a clade, but their circumscription differs from the traditional views. The subgenera recognized in Luzula remain mainly unresolved. A well-supported clade is represented by an assemblage of representatives of five genera and species distributed in the Southern Hemisphere: Juncus capensis and J. lomatophyllus (both from section Graminifolii), Rostkovia, Distichia, Marsippospermum, and Patosia.
Similar content being viewed by others
Author information
Authors and Affiliations
Corresponding author
Additional information
The main part of this study was supported by EU grants: SYS-RESOURCE (The Natural History Museum, London, Jodrell Laboratory, RBG Kew, UK) and COBICE (Institute of Botany, University of Copenhagen, DK), where samples were collected and some DNA isolation, PCR reactions and sequencing were performed. L. D. thanks Mark W. Chase for his great hospitality during the period spent at Jodrell Laboratory, RBG Kew and curator of the Herbarium where many Juncaceae DNA samples were collected. Charlotte Hansen and Lisbeth Knudsen (both University of Copenhagen) provided skilful technical assistance. Part of the study was done in the DNA Laboratory, Institute of Botany ASCR and Laboratory of the Centre for Integrated Genomics and Institute of Molecular Genetics ASCR and was supported in part by grants GACR 206/02/035, GAUK 109/2002/B/BIO and AVOZ 6005908. We also would like to thank two anonymous reviewers for comments on the manuscript.
Rights and permissions
About this article
Cite this article
Drábková, L., Kirschner, J., Seberg, O. et al. Phylogeny of the Juncaceae based on rbcL sequences, with special emphasis on Luzula DC. and Juncus L.. Plant Syst. Evol. 240, 133–147 (2003). https://doi.org/10.1007/s00606-003-0001-6
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-003-0001-6