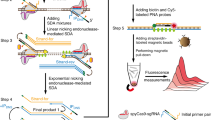
Abstract
A probing system has been developed based on dual-site ligation-assisted loop-mediated isothermal amplification (dLig-LAMP) for the selective colorimetric detection of SARS-CoV-2. This approach can induce false-positive and -negative detection in real clinical samples; dLig-LAMP operates with improved selectivity. Unlike RT-LAMP, the selectivity of dLig-LAMP is determined in both the ligation and primer binding steps, not in the reverse transcription step. With this selective system in hand, we developed a colorimetric signaling system for point-of-care detection. We also developed a colorimetric probe for sensing pyrophosphate, which arises as a side product during the LAMP DNA amplification. Thus, dLig-LAMP appears to be an alternative method for improving the selectivity problems associated with reverse transcription. In addition, combining dLig-LAMP with colorimetric pyrophosphate probing allows point-of-care detection of SARS-CoV-2 within 1 h with high selectivity.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Viruses are pernicious because they can spread and infect their targets rapidly [1]. Many dangerous viruses have their own RNA genomes that readily mutate [2]. Novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a new type of coronavirus (SARS-CoV appeared in 2002–2004; MERS-CoV in 2015 [3, 4]) that was first detected at the end of 2019 and remains spreading to this day [5, 6]. Such viruses that are highly infectious can result in pandemics [7]. Therefore, their rapid diagnosis of viruses is vital.
The reverse transcription-based polymerase chain reaction (RT-PCR) is commonly used for the detection of viral RNA because it has proven its ability to provide sensitive and selective detection of viral genomes [8, 9]. Nevertheless, it has several drawbacks in point-of-care detection: namely, it is time-consuming and demands expensive instrumentation [10]. Thus, many alternative methods have been developed for simple and rapid point-of-care detection [11, 12]. For the purpose of viral RNA detection, new techniques for rapid isothermal amplification, which is pivotal in rapid point-of-care detection, have been realized using, for example, recombinase polymerase amplification (RPA) [13,14,15], rolling circle amplification (RCA) [16, 17], and loop-mediated isothermal amplification (LAMP) [18, 19]. Nevertheless, some of isothermal amplification-based point-of-care detection systems have a limitation because they operate with low selectivity when compared with the RT-PCR method for the diagnosis of viral RNA [20,21,22]; the RT-PCR method itself also exhibits the same selectivity problem, because of mispairing or self-pairing of the primer during the reverse transcription targeting viral RNA, but recently this problem has been overcome in several ways [23, 24]. Point-of-care detection still requires the issue of selectivity to be addressed when using isothermal amplification methods.
In this study, we wished to modify the general LAMP system as an isothermal amplification method for the detection of SARS-CoV-2, because it allows rapid amplification with sufficient sensitivity. Typically, the LAMP system requires reverse transcriptase (RT) to produce target cDNA using a primer sequence that binds to viral RNA. Unfortunately, during the process of reverse transcription using this primer, many erroneous forms of cDNA are produced.
The first problem is the possibility of false-positive diagnosis [25,26,27]. When extracting the actual target viral RNA, the possibility of the existence of other similar viral RNAs, and the possibility that some sequences in the human genome are similar, is implied. In this case, it can be difficult for the primer to distinguish among them; furthermore, there is the possibility of false-positive detection even if there is no actual target RNA. Therefore, it is important to design a primer with a target that does not overlap with areas any other. Thus, most primers have targeted a conservative site, like the nucleoprotein, during RT-LAMP. If the primer sequence is too specific, however, it tends to become difficult to bind to the target RNA, thereby leading to the possibility of false-negatives. That is, even if the actual target RNA is present, there may be cases where the primer cannot bind to it and, thus, the target cDNA cannot be formed. The problem of false-positives may be solved by allowing the cDNA to be formed only under restricted conditions, when everything is perfectly combined. The problem of false-negatives may be solved by increasing the length of the combined sequence.
To overcome the selectivity problems of LAMP, in this study we developed a unique dual-site ligation-assisted cDNA synthesis—without using reverse transcriptase—for application to the LAMP process. Here, we used Splint R Ligase, an RNA-templated DNA ligase, that has a reaction rate much faster than that of T4 DNA Ligase [28]. We employed Splint R Ligase with three DNA oligonucleotide templates that are complementary to the specific region of the target RNA sequence. Ligation of the three DNA oligonucleotides using Splint R Ligase could produce the cDNA within several minutes, but only in the presence of the target RNA, and this cDNA was then amplified by the LAMP reaction system. Our dual-site ligation-assisted LAMP (dLig-LAMP) reaction requires the two ligation sites to produce the cDNA, which would not be synthesized if there were any mismatched sequences or if either of the ligation templates was not bound, because there would be no ligation event. Furthermore, one of the DNA oligonucleotide templates was sufficiently long (76–77mer) to bind to the target RNA. Finally, if no cDNA were formed it could not be amplified. Thus, we believe that this dLig-LAMP system is a promising tool for solving the selectivity problems that arise when using isothermal amplification methods.
Colorimetric sensing is useful for point-of-care detection because it allows simple and convenient detection by the naked eye, without the need for any detection instruments. Thus, we have also developed a colorimetric pyrophosphate-sensing probe (PP Probe) because, during isothermal DNA amplification, a large amount of pyrophosphate is produced as a side product [29, 30]. We then combined our PP Probe with the dLig-LAMP system for the colorimetric detection of SARS-CoV-2. It was difficult to apply our PP Probe to RT-LAMP because dithiothreitol (DTT) is essential to maintain the activity of reverse transcriptase during the reverse transcription process, and DTT is also sensed by the PP Probe. Gratifyingly, the dLig-LAMP system eliminates this step, thereby allowing the PP Probe to selectively detect amplified pyrophosphate only in the presence of the target RNA. Therefore, our dLig-LAMP system combined with the PP Probe allowed the rapid, selective, and colorimetric point-of-care detection of SARS-CoV-2.
Using this newly developed diagnostic method, we conducted a clinical diagnostic experiment with a patient infected with COVID-19. Detection was possible with high accuracy in terms of a rapid color change. Although this completely new concept in isothermal genetic diagnosis requires further verification, it appears to minimize the effort required for primer design while offering new possibilities in detection.
Experiments
General information
All DNA oligonucleotides and the dNTP mixture (dATP, dTTP, dCTP, dGTP) were purchased from Bioneer and Cosmo Genetech (South Korea). Target RNA and mismatched target RNA were synthesized using in vitro transcription. Splint R Ligase, WarmStart RTx Reverse Transcriptase, and Bst 2.0 WarmStart DNA polymerase were obtained from New England Biolabs (USA). The PP Probe was prepared according to a previously reported procedure [29, 30]; its spectra were in accordance with those described. UV–Vis absorption spectra were recorded using a Shimadzu (Japan) UV-1650PC spectrophotometer. Fluorescence was recorded using the PF-6500 spectrofluorometer (JASCO, Japan). All optical measurements were performed at room temperature, using a quartz cuvette (path length: 1 cm).
All gel electrophoresis was performed in 20% polyacrylamide gel (PAGE). Forty-percent acrylamide/Bis solution 29:1 (purchased from Bio-Rad, USA; 15 mL), 10 × TBE buffer (3 mL), and 20% ammonium persulfate solution (dissolved in H2O; 300 mL) were mixed in one tube and water was added to a total volume of 30 mL. TEMED was added to make 20% polyacrylamide gel. The gels were loaded in an electrophoresis instrument (CBS Scientific, CA, USA) and treated at 180 V for 14 h. The gels were stained in EtBr solution for 10 min; the stained gels were washed in water for 30 min. The gel photos and colorimetric detection images were captured by a mobile device under a transilluminator.
dLig-LAMP reaction
The dLig-LAMP reaction was performed with a total solution volume of 20 μL. The LAMP primer mixture was prepared including 16 μM of FIP/BIP primers, 2 μM of F3/B3 primers, and 4 μM of LF/LB primers. The cDNA template mixture was prepared including 10 nM of the LT-1/LT-2/LT-3 templates. For one dLig-LAMP reaction, the LAMP primer mixture (2 μL), the cDNA template mixture (2 μL), 10 × isothermal amplification buffer [200 mM of Tris–HCl, 100 mM of (NH4)2SO4, 500 mM of KCl, 20 mM of MgSO4; pH 8.8 at 25 °C; 2 μL], 10 × Splint R Ligase buffer (500 mM of Tris–HCl, 100 mM of MgCl2, and 10 mM of ATP; pH 7.5 at 25 °C; 2 μL), and dNTP mixture (2 mM of dATP, dCTP, dGTP, and dTTP; 5 μL) were added into a 1.5-mL tube. The target (5 μL) was added. Finally, the enzymes Splint R Ligase (25 U/μL; 1 μL) and Bst 2.0 WarmStart DNA polymerase (8 U/μL; 1 μL) were added into the reaction. The mixture was incubated for 15 min at 37 °C and then for 45 min at 65 °C.
Primer and template negative controls of dLig-LAMP
For the negative control studies, the dLig-LAMP reactions were performed using the standard procedure, but with one of the primer or templates removed. All reactions were monitored using PAGE. The colorimetric detection buffer (30% of 10 mM HEPES buffer and 70% acetonitrile; 180 μL) was added into the dLig-LAMP mixture to give a total volume of 200 μL for the colorimetric detection assay. The PP Probe (25 mM, 1 μL) was added into the reaction tube, which was then shaken for 1 min. For detailed analysis, absorbances were measured of the reactions performed in the presence of the PP Probe.
Sensitivity and selectivity measurements
The dLig-LAMP reactions were performed using our standard procedure. For sensitivity measurements, solutions of the target RNA were prepared with concentrations ranging from 1 aM to 1 nM. Reactions were performed three times to determine the reproducibility. The sensitivity was measured in terms of absorbance. For selectivity measurements, three different targets (matched target, one-base-mismatched target, and two-base-mismatched target) were used. The PAGE results were compared with those obtained using the RT-LAMP assay. One RT-LAMP reaction included the LAMP primer mixture (2 μL), 10 × isothermal amplification buffer [200 mM of Tris–HCl, 100 mM of (NH4)2SO4, 500 mM of KCl, 20 mM of MgSO4; pH 8.8 at 25 °C; 2 μL], the dNTP mixture (2 mM of dATP, dCTP, dGTP, and dTTP; 5 μL), water (4 μL), the target (5 μL), WarmStart RTx Reverse Transcriptase (15 U/μL; 1 μL), and Bst 2.0 WarmStart DNA polymerase (8 U/μL; 1 μL). The RT-LAMP reaction mixture was incubated at 65 °C for 1 h. Each reaction was performed three times to determine the reproducibility of the dLig-LAMP assays. The selectivity was measured in terms of absorbance.
Multiple ligation-assisted LAMP reaction
We prepared the short-ligation template mixture to include 10 nM of LTs-1, LTs-2, LTs-3, LTs-4, LTs-5, LTs-6, LTs-7, LTs-8, LTs-9, LTs-10, and LTs-11. For comparison, we prepared one template-mismatched short-ligation template mixture, including 10 nM of LTs-1, LTs-2, LTs-3, LTs-4, LTs-5, LTs-6 mismatch, LTs-7, LTs-8, LTs-9, LTs-10, and LTs-11. For one multiple ligation-assisted LAMP reaction, we added the LAMP primer mixture (2 μL) and the short-ligation template mixture or one template-mismatched short-ligation template mixture (2 μL); all of the other protocols were the same as those for the dLig-LAMP process. The target RNA concentration was 1 nM. All reactions were confirmed using PAGE. The colorimetric detection buffer (30% of 10 mM HEPES buffer and 70% acetonitrile; 180 μL) was added into the dLig-LAMP reaction mixture to give a total volume of 200 μL for the colorimetric detection assay. The PP Probe (25 mM, 1 μL) was added into the reaction tube, which was then shaken for 1 min. For detailed analysis, absorbances were measured of the reactions performed in the presence of PP Probe.
Full-genome SARS-CoV-2 sensitivity and bacteria genome selectivity study
An AccuPlex™ SARS-CoV-2 Reference Material Kit (Seracare, Milford, MA, USA), which was assigned as 5000 copies/mL, was used for spiked samples. SARS-CoV-2 RNA was extracted using eMAG (BioMerieux, MarcylEtoile, France), following the extraction protocol provided by the manufacturer, with an input volume of 200 μL and an elution volume of 50 μL. The copy concentration in the extracted RNA would be approximately 20 copies/μL. To increase the concentration, the SARS-CoV-2 RNA was lyophilized to give a concentration of 200 copies/μL. For the sensitivity study, the RNA sample was diluted in water to give concentrations ranging from 1.6 to 200 copies/μL. The absorbances of all samples were measured using the dLig-LAMP system and the PP Probe; a linear plot was calculated to obtain the LOD. For the selectivity study, samples of nine types of bacterial genomes, which are normal flora in the upper respiratory tract, were prepared. All samples of bacterial DNA were extracted by the boiling method using DNA extraction buffer (Seegene, Seoul, South Korea). The extracted bacterial genomes were tested using the dLig-LAMP assay and the PP Probe and compared with the SARS-CoV-2 genome. For detailed analysis, absorbances and fluorescence were measured of the reactions performed in the presence of the PP Probe.
Clinical sample preparation and validation
This study was approved by the Jeonbuk National University Hospital Institutional Review Board (CUIH 2021–11-005). A total of 40 residual samples for SARS-CoV-2 and 5 other virus samples (Influenza A Virus, Influenza B Virus, Respiratory Syncytial Virus A, Respiratory Syncytial Virus B, and Human Rhinovirus) real-time reverse-transcription PCR (rRT-PCR) were enrolled in this study: 20 positive samples, 20 negative samples, and 5 other virus samples. The samples were obtained from a nasopharyngeal swab collected in an eNAT tube (Copan Italy, Brescia, Italy); they were stored at − 20 °C after clinical tests. The nucleic acid was extracted with Magna Pure 24 (Roche Diagnostics, Basel, Swiss) or eMAG (BioMérieux, Marcy-l’Étoile, France), using the manufacturer’s protocol. The rRT-PCR tests were performed using the Allplex SARS-CoV-2 Assay (Seegene, Seoul, South Korea); the cycle threshold (Ct) values of the positive samples ranged from 22.33 to 36.15 in the N gene. For clinical validation, 40 reactions for SARS-CoV-2 tests and 6 reactions for selectivity tests were performed using our dLig-LAMP reaction with the PP Probe. For detailed analysis, absorbances were measured of the reactions performed in the presence of the PP Probe.
Results and discussion
Optimization
Based on our hypothesis, we designed three templates (LT1, LT2, LT3) for the synthesis of target cDNA mediated by Splint R Ligase (Table S1). Among them, the LT2 and LT3 templates presented a monophosphate unit at the 5′-end for the ligation event. The templates containing a phosphate modification at the 5′-end would also allow the ligation by Splint R Ligase to occur in the presence of perfectly matched target RNA. We designed six primers (F3, B3, FIP, BIP, LF, LB; Table S1) for the LAMP amplification process. To examine the selectivity, we designed not only the target RNA but also one- and two-base-mismatched target RNAs (Table S1). We highlight the mismatched points in the target RNA with red characters in Scheme 1 (black box). Theoretically, a general RT-PCR or RT-LAMP system cannot discriminate the perfectly matched sequence from one- or two-base-mismatched sequences because when the reaction primers bind to the target RNA, they might be able to amplify the gene with mismatched-sequence amplicons. In contrast, we expected that our dLig-LAMP system might discriminate mismatched points during the ligation step. Splint R Ligase produces complementary cDNA sequences only in the presence of a perfectly matched target RNA sequence; if one of the ligation templates does not match, the cDNA would not be synthesized. Therefore, we expected that the selectivity of our dLig-LAMP assay would be superior to that of general RT-LAMP or RT-PCR.
(blue box) Schematic representation of dLig-LAMP and RT-LAMP assays. (pink box) Mechanism of PP Probe sensing through the recognition of pyrophosphate. (black box) LAMP primer binding sites (marked in blue), model mismatch sequence sites (marked in red), and ligation sites (marked in green) in the SARS-CoV-2 N gene sequence (GenBank NC_045512.2)
First, we confirmed that the dLig-LAMP assay would operate in the presence of the SARS-CoV-2 N gene sequence. Figure 1a provides clear evidence that our dLig-LAMP assay functioned for the target SARS-CoV-2 N gene sequence, but not in the absence of any of the primers or templates. The dLig-LAMP system requires six primers and three templates. According to our polyacrylamide gel electrophoresis (PAGE) image, the negative control lanes (lanes 1–7) did not exhibit any amplification patterns, but lane 8, in which all of the primers and templates were present, provided the amplification pattern. Thus, dLig-LAMP operated with all of the primers for LAMP and with the three templates, but not at all when one of them was absent. Next, we examined the possibility of colorimetric detection when using the PP Probe to sense the pyrophosphate formed during DNA amplification (Fig. 1b). The dLig-LAMP assay released an abundance of pyrophosphate during the LAMP DNA amplification; the released pyrophosphate could extract the Cu2+ ion from the PP probe, resulting in a color change from pink to colorless. We observed such a dramatic color change only in the presence of the SARS-CoV-2 N gene; similar to the PAGE observations, none of the other samples 1–7 resulted in any color change. Absorption spectra revealed a dramatic decrease in absorption at 555 nm (λmax), the characteristic signal of the PP Probe, in the presence of the target SARS-CoV-2 N gene (sample 8), but no such changes for any of the other samples (Fig. 1c). Thus, combining the dLig-LAMP system with the PP Probe allowed the detection of the target SARS-CoV-2 N gene with a colorimetric signal change.
Primer- and template-free negative control experiments using dLig-LAMP. Sample 1: dLig-LAMP without LT-1, sample 2: dLig-LAMP without LT-2, sample 3: dLig-LAMP without LT-3, sample 4: dLig-LAMP without F3, sample 5: dLig-LAMP without B3, sample 6: dLig-LAMP without FIP, sample 7: dLig-LAMP without BIP, and sample 8: dLig-LAMP (positive control). All dLig-LAMP reaction mixtures included 1 nM of target RNA. (a) Results of 20% PAGE: lane m: DNA marker; lanes 1–8 correspond to samples 1–8. (b) Colorimetric detection assay of sample 1–8 using PP Probe. (c) Absorbance spectra of samples 1–8 when using PP Probe
Sensitivity and selectivity with model target
Next, we measured the sensitivity of our dLig-LAMP assay when varying the concentration of the target RNA from 1 aM to 1 nM. Figure 2a presents the absorbance spectra of the PP Probe recorded after performing the dLig-LAMP reaction at the various concentrations. The spectra reveal that the sensitivity of the dLig-LAMP assay depended on the log concentration of the target RNA. To obtain the exact sensitivity, we measured the limit of detection (LOD) from three repeated concentration-dependent experiments. From the linear plot of the average absorbance change value obtained at each concentration (Fig. 2b), we calculated the LOD value using the 3σ method [LOD = 3 × (SD/S), where SD is the standard deviation and S is the slope of the logarithmic plot]. The LOD when using the dLig-LAMP assay was 1.36 fM.
Sensitivity of the dLig-LAMP system. (a) Absorption spectra of the dLig-LAMP reaction mixtures obtained in the presence of the target RNA at concentrations ranging from 10 nM to 1 aM, and in the absence of the target RNA. (b) Linear relationship between the absorbance at 555 nm and the logarithm of the concentration of the target RNA. A0 absorbance in the absence of target, A absorbance in the presence of target y-axis absolute value. Each point represents three repeated measurements, with error bar
The selectivity of the dLig-LAMP system was much higher than that of RT-LAMP system, as determined using PAGE (Fig. 3a). The dLig-LAMP assay clearly discriminated the perfectly matched sequence from one- to two-base-mismatched sequences, with the appearance of an amplification band. In contrast, the RT-LAMP assay did discriminate between the perfectly matched and mismatched sequences; all of the sequences, except for the sample obtained without the target, provided the band corresponding to the amplified DNA. We suspect that the mismatched sequences also bound to the SARS-CoV-2 N gene and underwent reverse transcription to produce cDNA. The dLig-LAMP assay did not produce the cDNA if any mismatches appeared in the template; without cDNA, the LAMP process could not function. When we added PP Probe in the dLig-LAMP assay system, we observed a dramatic color change, from pink to colorless, only in the presence of the target SARS-CoV-2 N gene (Fig. 3b). Figure 3c displays the absorbance intensities of the dLig-LAMP/PP Probe assays performed with the SARS-CoV-2 N gene, the one-base-mismatched target, and the two-base-mismatched target, and without the target. The dLig-LAMP assay was highly selective in discriminating the perfectly matched target sequence from the mismatched sequences, even with only one base mismatched. Although the selectivity of the dLig-LAMP system was higher than that of the general RT-LAMP system, we suspected that it might be difficult to distinguish a mismatch if it appeared at an irregular position, due to the limited length of the ligation template. In other words, if the template were shortened, the stability of the duplex would likely be sensitive to differences in the perfectly matched and mismatched sequences, and it would sufficiently distinguish mismatches from the perfectly matched sequence in all regions.
Selectivity of the dLig-LAMP assay. (a) PAGE of the dLig-LAMP assay (right gel image) and the RT-LAMP assay (left gel image). Lane m DNA marker, lane 1 reaction with target, lane 2 reaction with one-base-mismatched target, lane 3 reaction with two-base-mismatched target, lane 4 reaction without target. (b) Colorimetric detection using dLig-LAMP and the PP Probe. (c) Absorbance (λmax = 555 nm) of the PP Probe in each dLig-LAMP reaction. Error bars represent by the results of three repeated measurements
Therefore, we examined the multiple ligation-assisted LAMP reaction using short-ligation templates (Figure S1). We designed 11 short-ligation templates (LTs-1 to LTs-11), with each LTs sequence being complementary to the target RNA (Table S2). In addition, we modified one template (LTs-6 mismatch) to contain a mismatched sequence, and again examined the selectivity when any of the templates were not matched with the target. Although the multiple ligation-assisted LAMP reaction also operated well, it would not proceed if one of the ligation templates were not a perfect match. This method might be useful for increasing the selectivity of the detection of viruses with sufficiently low rates of false-positives.
Sensitivity and selectivity with SARS-CoV-2 and bacteria
To examine the practical applications of this system, we used the full genome of SARS-CoV-2 to perform a sensitivity study (Fig. 4a). We measured the absorption spectra in three repeated experiments while varying the number of copies of the SARS-CoV-2 full genome from 8 to 1000 copies/rxn; we obtained an LOD (3σ method) of 61.4 copies/rxn. Corman reported 2.31 log copies/mL based on a 25 μL reaction volume [9], and Pfefferle reported 2.83 log copies/mL of limit of detection in the SARS-CoV-2 real-time RT-PCR [31]. Even though our detection limit is lower than the RT-PCR system, it has strong points than RT-PCR such as detection time (our system needs 1 h totally), heavy instrument is not required, and simple treatment. Thus, the LOD when using our colorimetric dLig-LAMP system appears to be applicable for point-of-care detection of SARS-CoV-2. In addition, the PP probe has fluorescence when the Cu2+ ion is lost. Thus, we tried to calculate the LOD value of the fluorometric assay (Figure S5). Fluorometric LOD value was 69.2 copies/rxn; it has a similar capability with absorbance assay. Therefore, our system can be used in both absorbance and fluorescence.
(a) Sensitivity of dLig-LAMP assays performed using full-genome SARS-CoV-2 at concentrations ranging from 8 to 1000 copies/reaction (copies/rxn). Plot of the absorbance at 555 nm with respect to the number of copies of SARS-CoV-2. Inset: linear relationship from 8 to 250 copies/rxn. A0 absorbance in the absence of target, A absorbance in the presence of target, y-axis absolute value. (b) Selectivity of the dLig-LAMP assay toward full-genome SARS-CoV-2, relative to various bacterial genomes, in terms of absorbance at 555 nm. All reactions were repeated three times to obtain error bar ranges
We examined the possibility of cross reactions when using nine bacterial genomes (Fig. 4b) that can be infect humans and are located in mucous membranes of mouth and nose [32,33,34]. It would be possible for real clinical samples to include these bacterial genomes, potentially inducing false-positive signals; if there were any cross reactions, SARS-CoV-2 would not be detected selectively. Thus, we examined the selectivity of the colorimetric dLig-LAMP system targeting SARS-CoV-2 when these bacteria were present. None of the nine bacteria reacted with our dLig-LAMP system in triply repeated experiments, with no pyrophosphate being produced. Thus, our system appears suitable for use as a point-of-care SARS-CoV-2 detection tool.
Clinical validation
Finally, we attempted to diagnose real clinical SARS-CoV-2 samples using our dLig-LAMP system with the PP Probe (Fig. 5). We prepared 20 positive samples and 20 negative samples from a patient. Many false-negative cases have been reported when using RT-LAMP to diagnose COVID-19 patients [35,36,37]. In contrast, our dLig-LAMP system provided 100% true-negatives through both colorimetric detection and absorbance spectral measurements. Furthermore, even the positive samples provided 95% true-positives selectively. Under the 30 Ct value, the clinical samples exhibited a strong signal change (complete color change); above the 30 Ct value, the clinical samples underwent less of a signal change when compared with the less-than-30 Ct value samples. Additionally, we confirmed the selectivity in the case of other types of the clinical virus genome. We have prepared five different viruses’ clinical samples (Influenza A Virus, Influenza B Virus, Respiratory Syncytial Virus A, Respiratory Syncytial Virus B, and Human Rhinovirus) and tested with SARS-CoV-2 (Figure S8). Its results showed that all viral genomes other than SARS-CoV-2 did not react with the dLig-LAMP/PP probe system. According to this clinical validation experiment, the selectivity of our dLig-LAMP assays was improved relative to that of the general RT-LAMP detection assay. We believe that our dual-site ligation route to the production of cDNA might be promising to use in some diagnoses that require greater selectivity and that it might also be suitable for point-of-care detection for diverse RNA-based diagnoses, because the entire procedure involves a simple and rapid (1 h) one-pot reaction.
Clinical validation of the dLig-LAMP/PP Probe system. Both color imaging and absorbance spectra were performed using 40 clinical samples (20 positive; 20 negative). (a) Colorimetric detection assays. Positive samples appear as weakly pink or colorless; negative samples appear pink in color. (b) Absorbance detection at 555 nm, presented as a bar graph. Positive samples are presented in red (P01–P20); negative samples in blue color (N01–N20). RT-PCR Ct values of positive samples are presented in parenthesis next to each sample number. Black dotted line represents the negative line
Conclusion
We have developed a novel dLig-LAMP assay to improve the selectivity of RNA detection. In general RNA detection using the LAMP system, reverse transcriptase is required to produce a cDNA. According to a concentration-dependent study using model target RNA, the LOD was 1.36 fM. Furthermore, the dLig-LAMP assay displayed high selectivity, discriminating even one- and two-base-mismatched targets. In a study using full-genome SARS-CoV-2, the dLig-LAMP system provided an LOD of 61.4 copies/rxn and LOD of 69.2 copies/rxn for the fluorometric assay, with no cross-reactions occurring in the presence of various bacteria. From a study of its clinical validation, our dLig-LAMP system displayed perfect selectivity, with 100% true-negative sample detection, and sufficient sensitivity for positive samples displaying 95% true detection. Also, we demonstrate the clear clinical selectivity using the several types of viruses. Therefore, we believe that our dLig-LAMP system might be useful for improving the selectivity problems associated with reverse transcription.
References
Goyal M, Tewatia N, Vashisht H, Jain R, Kumar S (2021) Novel corona virus (COVID-19); global efforts and effective investigational medicines: a review. J Infect Public Health 14:910–921
Sanjuán R, Domingo-Calap P (2016) Mechanisms of viral mutation. Cell Mol Life Sci 73:4433–4448
Kirchdoerfer RN, Ward AB (2019) Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun 10:2342
da Costa VG, Moreli ML, Saivish MV (2020) The emergence of SARS, MERS and novel SARS-2 coronaviruses in the 21st century. Arch Virol 165:1517–1526
Carlos WG, Dela Cruz CS, Cao B, Pasnick S, Jamil S (2020) Novel Wuhan (2019-nCoV) coronavirus. Am J Respir Crit Care Med 201:P7–P8
Thompson R (2020) Pandemic potential of 2019-nCoV. Lancet Infect Dis 20:280
Amato A, Caggiano M, Amato M, Moccia G, Capunzo M et al (2020) Infection control in dental practice during the COVID-19 pandemic. Int J Environ Res Public Health 17:4769
Binnicker MJ (2020) Emergence of a novel coronavirus disease (COVID-19) and the importance of diagnostic testing: why partnership between clinical laboratories, public health agencies, and industry is essential to control the outbreak. Clin Chem 66:664–666
Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A et al (2020) Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 25:2000045
Brundha MP, Ezhilarasan D (2020) Advantages and disadvantages of RT-PCR in COVID 19. EJMCM 7:1174–1181
Suea-Ngam A, Bezinge L, Mateescu B, Howes PD, deMello AJ et al (2020) Enzyme-assisted nucleic acid detection for infectious disease diagnostics: moving toward the point-of-care. ACS Sens 5:2701–2723
Zhang Z, Ma P, Ahmed R, Wang J, Akin D et al (2022) Advanced point-of-care testing technologies for human acute respiratory virus detection. Adv Mater 34:2103646
Patchsung M, Jantarug K, Pattama A, Aphicho K, Suraritdechachai S et al (2020) Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng 4:1140–1149
Choi MH, Lee J, Seo YJ (2021) Combined recombinase polymerase amplification/rkDNA–graphene oxide probing system for detection of SARS-CoV-2. Anal Chim Acta 1158:338390
El Wahed AA, Patel P, Maier M, Pietsch C, Rüster D et al (2021) Suitcase lab for rapid detection of SARS-CoV-2 based on recombinase polymerase amplification assay. Anal Chem 93:2627–2634
Chaibun T, Puenpa J, Ngamdee T, Boonapatcharoen N, Athamanolap P et al (2021) Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat Commun 12:802
Jiao J, Duan C, Xue L, Liu Y, Sun W et al (2020) DNA nanoscaffold-based SARS-CoV-2 detection for COVID-19 diagnosis. Biosens Bioelectron 167:112479
Huang WE, Lim B, Hsu CC, Xiong D, Wu W et al (2020) RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb Biotechnol 13:950–961
Broughton JP, Deng X, Yu G, Fasching CL, Servellita V et al (2020) CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol 38:870–874
Karami A, Gill P, Kalantar Motamedi MH, Saghafinia MJ (2011) A review of the current isothermal amplification techniques: applications, advantages and disadvantages. Glob Infect Dis 3:293–302
Daher RK, Stewart G, Boissinot M, Boudreau DK, Bergeron MG (2015) Influence of sequence mismatches on the specificity of recombinase polymerase amplification technology. Mol Cell Probes 29:116–121
Zhang SQ, Tan B, Li P, Wang FX, Guo L et al (2014) Comparison of conventional RT-PCR, reverse-transcription loop-mediated isothermal amplification, and SYBR green I-based real-time RT-PCR in the rapid detection of bovine viral diarrhea virus nucleotide in contaminated commercial bovine sera batches. J Virol Methods 207:204–209
Seibel P, Degoul F, Romero N, Marsac C, Kadenbach B (1990) Identification of point mutations by mispairing PCR as exemplified in MERRF disease. Biochem Biophys Res Commun 173:561–565
Lin FH, Lin R (1992) A comparison of single nucleotide primer extension with mispairing PCR-RFLP in detecting a point mutation. Biochem Biophys Res Commun 189:1202–1206
Chou PH, Lin YC, Teng PH, Chen CL, Lee PY (2011) Real-time target-specific detection of loop-mediated isothermal amplification for white spot syndrome virus using fluorescence energy transfer-based probes. J virol methods 173:67–74
Hsieh K, Mage PL, Csordas AT, Eisenstein M, Soh HT (2014) Simultaneous elimination of carryover contamination and detection of DNA with uracil-DNA-glycosylase-supplemented loop-mediated isothermal amplification (UDG-LAMP). Chem commun 50:3747–3749
Stahlberg A, Hakansson J, Xian X, Semb H, Kubista M (2004) Properties of the reverse transcription reaction in mRNA quantification. Clin chem 50:509–515
Jin J, Vaud S, Zhelkovsky AM, Posfai J, McReynolds LA (2016) Sensitive and specific miRNA detection method using SplintR Ligase. Nucleic Acids Res 44:e116–e116
Pandith A, Seo YJ (2020) Label-free sensing platform for miRNA-146a based on chromo-fluorogenic pyrophosphate recognition. J Inorg Biochem 203:110867
Choi MH, Seo YJ (2021) Rapid and highly sensitive hairpin structure-mediated colorimetric detection of miRNA. Anal Chim Acta 1176:338765
Pfefferle S, Reucher S, Nörz D, Lütgehetmann M (2020) Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Eurosurveillance 25:2000152
Dahlén G (2000) (2009) Bacterial infections of the oral mucosa. Periodontol 49:13–38
Lai CC, Chen SY, Ko WC, Hsueh PR (2021) Increased antimicrobial resistance during the COVID-19 pandemic. Int J Antimicrob Agents 57:106324
Song W, Jia X, Zhang X, Ling Y, Yi Z (2021) Co-infection in COVID-19, a cohort study. J Infect 82:414–451
Ganguli A, Mostafa A, Berger J, Lim J, Araud E et al (2021) Reverse transcription loop-mediated isothermal amplification assay for ultrasensitive detection of SARS-CoV-2 in saliva and viral transport medium clinical samples. Anal Chem 93:7797–7807
Mohon AN, Oberding L, Hundt J, van Marle G, Pabbaraju K et al (2020) Optimization and clinical validation of dual-target RT-LAMP for SARS-CoV-2. J Virol Methods 286:113972
Rödel J, Egerer R, Suleyman A, Sommer-Schmid B, Baier M et al (2020) Use of the variplex™ SARS-CoV-2 RT-LAMP as a rapid molecular assay to complement RT-PCR for COVID-19 diagnosis. J Clin Virol 132:104616
Funding
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (2021R1A2C1003804), funded by the Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
604_2022_5293_MOESM1_ESM.docx
Supplementary file1 (DOCX 1494 KB) Details of the oligonucleotide sequences; data for selectivity study by short ligation template; original UV absorption spectra.
Rights and permissions
About this article
Cite this article
Choi, M.H., Lee, J. & Seo, Y.J. Dual-site ligation-assisted loop-mediated isothermal amplification (dLig-LAMP) for colorimetric and point-of-care determination of real SARS-CoV-2. Microchim Acta 189, 176 (2022). https://doi.org/10.1007/s00604-022-05293-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-022-05293-7