Abstract
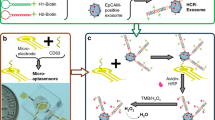
A novel surface plasmon resonance (SPR) strategy is introduced for the specific determination of exosomes based on aptamer recognition and polydopamine-functionalized gold nanoparticle (Au@PDA NP)–assisted signal amplification. Exosomes derived from hepatic carcinoma SMMC-7721 were selected as the model target. SMMC-7721 exosomes can be specifically captured by the aptamer ZY-sls that was complementary to the DNA tetrahedron probes (DTPs), and then the CD63 aptamer–linked Au@PDA NPs recognized SMMC-7721 exosomes for signal amplification. The DTPs were modified on a Au film for preventing Au deposition on the surface during the introduction of HAuCl4, and PDA coated on the AuNPs was used to reduce HAuCl4 in situ without any reductant assistance. It results in a further enhanced SPR signal. The assay can clearly distinguish SMMC-7721 exosomes from others (HepG2 exosomes, Bel-7404 exosomes, L02 exosomes, MCF-7 exosomes, and SW480 exosomes, respectively). SMMC-7721 exosomes are specifically determined as low as 5.6 × 105 particles mL−1. The method has successfully achieved specific determination of SMMC-7721 exosomes even in 50% of human serum without any pretreatment.

A novel surface plasmon resonance (SPR) strategy was introduced for the specific determination of exosomes based on aptamer recognition and polydopamine functionalized gold nanoparticles (Au@PDA NPs). The SPR signal was improved using the Au@PDA NPs assisted amplification.






Similar content being viewed by others
References
van Niel G, D’Angelo G, Raposo G (2018) Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19:213–228
Yang BW, Chen Y, Shi JL (2018) Exosome biochemistry and advanced nanotechnology for next-generation theranostic platforms. Adv Mater:31. https://doi.org/10.1002/adma.201802896
Brown L, Wolf JM, Prados-Rosales R, Casadevall A (2015) Through the wall: extracellular vesicles in gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol 13:620–630
Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H (2018) New technologies for analysis of extracellular vesicles. Chem Rev 118:1917–1950
Pick H, Alves AC, Vogel H (2018) Single-vesicle assays using liposomes and cell-derived vesicles: from modeling complex membrane processes to synthetic biology and biomedical applications. Chem Rev 118:8598–8654
Boriachek K, Masud MK, Palma C, Phan HP, Yamauchi Y, Hossain MSA, Nguyen NT, Salomon C, Shiddiky MJA (2019) Avoiding pre-isolation step in exosome analysis: direct isolation and sensitive detection of exosomes using gold-loaded nanoporous ferric oxide nanozymes. Anal Chem 91:3827–3834
Boriachek K, Islam MN, Gopalan V, Lam AK, Nguyen NT, Shiddiky MJA (2017) Quantum dot-based sensitive detection of disease specific exosome in serum. Analyst 142:2211–2220
Chen Z, Cheng SB, Cao P, Qiu QF, Chen Y, Xie M, Xu Y, Huang WH (2018) Detection of exosomes by ZnO nanowires coated three-dimensional scaffold chip device. Biosens Bioelectron 122:211–217
Kang YT, Kim YJ, Bu J, Cho YH, Han SW, Moon BI (2017) High-purity capture and release of circulating exosomes using an exosome-specific dual-patterned immunofiltration (ExoDIF) device. Nanoscale 9:13495–13505
Lewis JM, Vyas AD, Qiu Y, Messer KS, White R, Heller MJ (2018) Integrated analysis of exosomal protein biomarkers on alternating current electrokinetic chips enables rapid detection of pancreatic cancer in patient blood. ACS Nano 12:3311–3320
Li T, Zhang R, Chen H, Huang Z, Wang XH, Deng A, Kong J (2018) An ultrasensitive polydopamine bi-functionalized SERS immunoassay for exosome-based diagnosis and classification of pancreatic cancer. Chem Sci 9:5372–5382
Zong SF, Wang L, Chen C, Lu J, Zhu D, Zhang YZ, Wang ZY, Cui YP (2016) Facile detection of tumor-derived exosomes using magnetic nanobeads and SERS nanoprobes. Anal Methods 8:5001–5008
Liu C, Zeng X, An Z, Yang Y, Eisenbaum M, Gu X, Jornet JM, Dy GK, Reid ME, Gan Q, Wu Y (2018) Sensitive detection of exosomal proteins via a compact surface plasmon resonance biosensor for cancer diagnosis. ACS Sens 3:1471–1479
Qiu GY, Thakur A, Xu C, Ng SP, Lee Y, Wu CML (2019) Detection of glioma-derived exosomes with the biotinylated antibody-functionalized titanium nitride plasmonic biosensor. Adv Funct Mater 29(9). https://doi.org/10.1002/adfm.201806761
Liang K, Liu F, Fan J, Sun D, Liu C, Lyon CJ, Bernard DW, Li Y, Yokoi K, Katz MH, Koay EJ, Zhao Z, Hu Y (2017) Nanoplasmonic quantification of tumour-derived extracellular vesicles in plasma microsamples for diagnosis and treatment monitoring. Nat Bio Eng 1(4):1–11. https://doi.org/10.1038/s41551-016-0021
Picciolini S, Gualerzi A, Vanna R, Sguassero A, Gramatica F, Bedoni M, Masserini M, Morasso C (2018) Detection and characterization of different brain-derived subpopulations of plasma exosomes by surface plasmon resonance imaging. Anal Chem 90:8873–8880
Wang YM, Liu JW, Adkins GB, Shen W, Trinh MP, Duan LY, Jiang JH, Zhong WW (2017) Enhancement of the intrinsic peroxidase-like activity of graphitic carbon nitride nanosheets by ssDNAs and its application for detection of exosomes. Anal Chem 89:12327–12333
Wang S, Zhang L, Wan S, Cansiz S, Cui C, Liu Y, Cai R, Hong C, Teng I, Shi M, Wu Y, Dong Y, Tan W (2017) Aptasensor with expanded nucleotide using DNA nanotetrahedra for electrochemical detection of cancerous exosomes. ACS Nano 11:3943–3949
Huang L, Wang DB, Singh N, Yang F, Gu N, Zhang XE (2018) A dual-signal amplification platform for sensitive fluorescence biosensing of leukemia-derived exosomes. Nanoscale 10:20289–20295
Zhang HX, Wang ZH, Zhang QX, Wang F, Liu Y (2019) Ti3C2 MXenes nanosheets catalyzed highly efficient electrogenerated chemiluminescence biosensor for the detection of exosomes. Biosens Bioelectron 124-125:184–190
Wang Q, Zou L, Yang X, Liu X, Nie W, Zheng Y, Cheng Q, Wang K (2019) Direct quantification of cancerous exosomes via surface plasmon resonance with dual gold nanoparticle-assisted signal amplification. Biosens Bioelectron 135:129–136
Huang R, He L, Xia Y, Xu H, Liu C, Xie H, Wang S, Peng L, Liu Y, Liu Y, He N, Li Z (2019) A sensitive aptasensor based on a hemin/G-quadruplex assisted signal amplification strategy for electrochemical detection of gastric cancer exosomes. Small 15. https://doi.org/10.1002/smll.201900735
Tan Y, Guo Q, Xie Q, Wang K, Yuan B, Zhou Y, Liu J, Huang J, He X, Yang X, He C, Zhao X (2014) Single-walled carbon nanotubes (SWCNTs)-assisted cell-systematic evolution of ligands by exponential enrichment (cell-SELEX) for improving screening efficiency. Anal Chem 86:9466–9472
Nie W, Wang Q, Zou L, Zheng Y, Liu X, Yang X, Wang K (2018) Low-fouling surface plasmon resonance sensor for highly sensitive detection of microRNA in a complex matrix based on the DNA tetrahedron. Anal Chem 90:12584–12591
Ye DK, Zuo XL, Fan CH (2018) DNA nanotechnology-enabled interfacial engineering for biosensor development. Annu Rev Anal Chem 11:171–195
Huang YL, Mo S, Gao ZF, Chen JR, Lei JL, Luo HQ, Li NB (2017) Amperometric biosensor for microRNA based on the use of tetrahedral DNA nanostructure probes and guanine nanowire amplification. Microchim Acta 184:2597–2604
Hu W, He G, Zhang H, Wu X, Li J, Zhao Z, Qiao Y, Lu Z, Liu Y, Li CM (2014) Polydopamine-functionalization of graphene oxide to enable dual signal amplification for sensitive surface plasmon resonance imaging detection of biomarker. Anal Chem 86:4488–4493
Scarano S, Pascale E, Palladino P, Fratini E, Minunni M (2018) Determination of fermentable sugars in beer wort by gold nanoparticles@polydopamine: a layer-by-layer approach for localized surface plasmon resonance measurements at fixed wavelength. Talanta 183:24–32
Choi CK, Li J, Wei K, Xu YJ, Ho LW, Zhu M, To KK, Choi CH, Bian L (2015) A gold@polydopamine core-shell nanoprobe for long-term intracellular detection of microRNAs in differentiating stem cells. J Am Chem Soc 137:7337–7346
Zhang JJ, Mou L, Jiang XY (2018) Hydrogels incorporating Au@polydopamine nanoparticles: robust performance for optical sensing. Anal Chem 90:11423–11430
Ye Q, Zhou F, Liu WM (2011) Bioinspired catecholic chemistry for surface modification. Chem Soc Rev 40:4244–4258
Fang T, Lv HW, Lv GS, Li T, Wang CZ, Han Q, Yu LX, Su B, Guo LN, Huang SN, Cao D, Tang L, Tang SH, Wu MC, Yang W, Wang HY (2018) Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun 9:191. https://doi.org/10.1038/s41467-017-02583-0
Funding
This work was carried out within the framework of the National Natural Science Foundation of China (21675047, 21375034, and 21735002) and the National Science Foundation for Distinguished Young Scholars of Hunan Province (2016JJ1008).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 992 kb)
Rights and permissions
About this article
Cite this article
Liao, G., Liu, X., Yang, X. et al. Surface plasmon resonance assay for exosomes based on aptamer recognition and polydopamine-functionalized gold nanoparticles for signal amplification. Microchim Acta 187, 251 (2020). https://doi.org/10.1007/s00604-020-4183-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-020-4183-1