Abstract
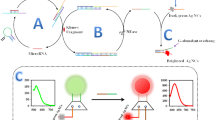
Using 6-carboxyfluorescein (FAM) and tetramethyl rhodamine (TAMRA) as fluorescent signals a ratiometric fluorescent three-dimensional (3D) DNA walker based on a catalytic hairpin assembly (CHA) reaction for microRNA-122 detection was constructed. This method uses CHA reaction triggered indirectly by the target to mediate the 3D DNA walker operation to amplify the signal. The dual emission ratio fluorescent signal with a single excitation wavelength was used as the signal output. This strategy combines DNA walker with CHA reaction and proportional fluorescence signal output methods, which can effectively reduce the background fluorescence signal and the risk of generating false-positive signals. Thus, the impact of environmental factors on the experiment is reduced, thereby obtaining reliable and stable experimental results. It uses the fluorescence excitation wavelength of 488 nm and the maximum fluorescence emission wavelength of 520 nm and 580 nm, respectively. It has a good linear response at a microRNA concentration range of 156.0 pM ~ 7.00 nM and a detection limit of 42.94 pM. This strategy has been successfully applied to detect microRNAs in spiked serum samples.

Schematic representation of three-dimensional (3D) DNA walker constructed using catalytic hairpin self-assembly reaction (CHA)-assisted amplification and ratiometric fluorescence signal output for the detection of miRNA-122 closely related to hepatitis





Similar content being viewed by others
References
Li D, Zhou W, Yuan R, Xiang Y (2017) A DNA-fueled and catalytic molecule machine lights up trace under-expressed microRNAs in living cells. Anal Chem 89(18):9934–9940. https://doi.org/10.1021/acs.analchem.7b02247
Yin H, Wang H, Li Z, Shu D, Guo P (2018) RNA micelles for the systemic delivery of anti-miRNA for cancer targeting and inhibition without ligand. ACS Nano 13(1):706–717. https://doi.org/10.1021/acsnano.8b07948
Ma P-Q, Liang C-P, Zhang H-H, Yin B-C, Ye B-C (2018) A highly integrated DNA nanomachine operating in living cells powered by an endogenous stimulus. Chem Sci 9(13):3299–3304. https://doi.org/10.1039/c8sc00049b
Tang Y, Liu M, Zhao Z, Li Q, Liang X, Tian J, Zhao S (2019) Fluorometric determination of microRNA-122 by using ExoIII-aided recycling amplification and polythymine induced formation of copper nanoparticles. Microchim Acta 186(3). https://doi.org/10.1007/s00604-019-3237-8
Li Q, Zhou S, Zhang T, Zheng B, Tang H (2020) Bioinspired sensor chip for detection of miRNA-21 based on photonic crystals assisted cyclic enzymatic amplification method. Biosens Bioelectron 150:111866. https://doi.org/10.1016/j.bios.2019.111866
Wang Y, Peng M, Wang W, Chen Y, He Z, Cao J, Lin Z, Yang Z, Gong M, Yin Y (2019) Verification of miRNAs in ginseng decoction by high-throughput sequencing and quantitative real-time PCR. Heliyon 5(4):e01418. https://doi.org/10.1016/j.heliyon.2019.e01418
Xiao M, Wang X, Li L, Pei H (2019) Stochastic RNA walkers for intracellular MicroRNA imaging. Anal Chem 91(17):11253–11258. https://doi.org/10.1021/acs.analchem.9b02265
Xie Y, Niu F, Yu A, Lai G (2019) Proximity binding-triggered assembly of two MNAzymes for catalyzed release of G-Quadruplex DNAzymes and an ultrasensitive homogeneous bioassay of platelet-derived growth factor. Anal Chem 92(1):593–598. https://doi.org/10.1021/acs.analchem.9b05002
Zhang Y, Chai Y, Wang H, Yuan R (2019) Target-induced 3D DNA network structure as a novel signal amplifier for ultrasensitive electrochemiluminescence detection of MicroRNAs. Anal Chem 91(22):14368–14374. https://doi.org/10.1021/acs.analchem.9b02817
Peng H, Newbigging AM, Reid MS, Uppal JS, Xu J, Zhang H, Le XC (2019) Signal amplification in living cells: a review of microRNA detection and imaging. Anal Chem 92(1):292–308. https://doi.org/10.1021/acs.analchem.9b04752
Zhou H, Liu J, Xu J-J, Zhang S-S, Chen H-Y (2018) Optical nano-biosensing interface via nucleic acid amplification strategy: construction and application. Chem Soc Rev 47(6):1996–2019. https://doi.org/10.1039/c7cs00573c
Ji D, Mou X, Kwok CK (2019) Label-free and ratiometric detection of microRNA based on target-induced catalytic hairpin assembly and two fluorescent dyes. Anal Methods 11(37):4808–4813. https://doi.org/10.1039/c9ay01891c
Li D, Yang F, Li X, Yuan R, Xiang Y (2020) Target-mediated base-mismatch initiation of a non-enzymatic signal amplification network for highly sensitive sensing of Hg2+. Analyst 145(2):507–512. https://doi.org/10.1039/c9an01836k
Mason SD, Tang Y, Li Y, Xie X, Li F (2018) Emerging bioanalytical applications of DNA walkers. TrAC Trends Anal Chem 107:212–221. https://doi.org/10.1016/j.trac.2018.08.015
Wei W, Wei M, Yin L, Pu Y, Liu S (2018) Improving the fluorometric determination of the cancer biomarker 8-hydroxy-2′-deoxyguanosine by using a 3D DNA nanomachine. Microchimica Acta 185(10):494. https://doi.org/10.1007/s00604-018-3036-7
Wang L, Wan Y, Xu Q, Lou X (2019) Long-term functional stability of functional nucleic acid–gold nanoparticle conjugates with different secondary structures. Langmuir 35(36):11791–11798. https://doi.org/10.1021/acs.langmuir.9b01884
Kang H, Buchman JT, Rodriguez RS, Ring HL, He J, Bantz KC, Haynes CL (2018) Stabilization of silver and gold nanoparticles: preservation and improvement of Plasmonic functionalities. Chem Rev 119(1):664–699. https://doi.org/10.1021/acs.chemrev.8b00341
Khan ZUH, Khan A, Chen Y, Shah NS, Muhammad N, Khan AU, Tahir K, Khan FU, Murtaza B, Hassan SU, Qaisrani SA, Wan P (2017) Biomedical applications of green synthesized Nobel metal nanoparticles. J Photochem Photobiol B Biol 173:150–164. https://doi.org/10.1016/j.jphotobiol.2017.05.034
Xiong E, Zhen D, Jiang L, Zhou X (2019) Binding-induced 3D-bipedal DNA walker for cascade signal amplification detection of thrombin combined with catalytic hairpin assembly strategy. Anal Chem 91(23):15317–15324. https://doi.org/10.1021/acs.analchem.9b04987
Jung C, Allen PB, Ellington AD (2015) A stochastic DNA walker that traverses a microparticle surface. Nat Nanotechnol 11(2):157–163. https://doi.org/10.1038/nnano.2015.246
Li W, Wang L, Jiang W (2017) A catalytic assembled enzyme-free three-dimensional DNA walker and its sensing application. Chem Commun 53(40):5527–5530. https://doi.org/10.1039/c7cc02306e
Ye T, Zhang Z, Yuan M, Cao H, Yin F, Wu X, Xu F (2020) An all-in-one Aptasensor integrating enzyme powered three-dimensional DNA machine for antibiotic detection. J Agric Food Chem 68(9):2826–2831. https://doi.org/10.1021/acs.jafc.9b08143
Wang J, Huang J, Quan K, Li J, Wu Y, Wei Q, Yang X, Wang K (2018) Hairpin-fuelled catalytic nanobeacons for amplified microRNA imaging in live cells. Chem Commun 54(73):10336–10339. https://doi.org/10.1039/c8cc06298f
Cheng Y, Dong L, Zhang J, Zhao Y, Li Z (2018) Recent advances in microRNA detection. Analyst 143(8):1758–1774. https://doi.org/10.1039/c7an02001e
Li D, Wu Y, Gan C, Yuan R, Xiang Y (2018) Bio-cleavable nanoprobes for target-triggered catalytic hairpin assembly amplification detection of microRNAs in live cancer cells. Nanoscale 10(37):17623–17628. https://doi.org/10.1039/c8nr05229h
Chen J, Yang H-H, Yin W, Zhang Y, Ma Y, Chen D, Xu Y, Liu S-Y, Zhang L, Dai Z, Zou X (2019) Metastable dumbbell probe-based hybridization chain reaction for sensitive and accurate imaging of intracellular-specific microRNAs in situ in living cells. Anal Chem 91(7):4625–4631. https://doi.org/10.1021/acs.analchem.8b05920
Yang Y, Huang J, Yang X, He X, Quan K, Xie N, Ou M, Wang K (2017) Gold nanoparticle based hairpin-locked-DNAzyme probe for amplified miRNA imaging in living cells. Anal Chem 89(11):5850–5856. https://doi.org/10.1021/acs.analchem.7b00174
Huang J, Zhu L, Ju H, Lei J (2019) Telomerase triggered DNA walker with a superhairpin structure for human telomerase activity sensing. Anal Chem 91(11):6981–6985. https://doi.org/10.1021/acs.analchem.9b01603
Sengupta D, Cassel T, K-y T, Aljuhani M, Chowdhary VK, Hu P, Zhang X, Fan TWM, Ghoshal K (2020) Regulation of hepatic glutamine metabolism by miR-122. Molecular Metabolism 34:174–186. https://doi.org/10.1016/j.molmet.2020.01.003
Grabar KC, Freeman RG, Hommer MB, Natan MJ (1995) Preparation and characterization of au colloid monolayers. Anal Chem 67(4):735–743. https://doi.org/10.1021/ac00100a008
Feng C, Wang Z, Chen T, Chen X, Mao D, Zhao J, Li G (2018) A dual-enzyme-assisted three-dimensional DNA walking machine using T4 polynucleotide kinase as activators and application in polynucleotide kinase assays. Anal Chem 90(4):2810–2815. https://doi.org/10.1021/acs.analchem.7b04924
Liu B, Liu J (2017) Freezing directed construction of bio/nano interfaces: reagentless conjugation, denser spherical nucleic acids, and better nanoflares. J Am Chem Soc 139(28):9471–9474. https://doi.org/10.1021/jacs.7b04885
Liu X, Wang L, Xu X, Zhao H, Jiang W (2018) Endogenous stimuli-responsive nucleus-targeted nanocarrier for intracellular mRNA imaging and drug delivery. ACS Appl Mater Interfaces 10(46):39524–39531. https://doi.org/10.1021/acsami.8b16345
Li X-L, Zhang Z-L, Zhao W, Xia X-H, Xu J-J, Chen H-Y (2016) Oriented assembly of invisible probes: towards single mRNA imaging in living cells. Chem Sci 7(5):3256–3263. https://doi.org/10.1039/c5sc04369g
Wang W, Ding X, Xu Q, Wang J, Wang L, Lou X (2016) Zeta-potential data reliability of gold nanoparticle biomolecular conjugates and its application in sensitive quantification of surface absorbed protein. Colloids Surf B: Biointerfaces 148:541–548. https://doi.org/10.1016/j.colsurfb.2016.09.021
Funding
This work was supported by the National Natural Science Foundation of China (no. 21465007, 21565007, 21165004, 21163002), the Guangxi Natural Science Foundation of China (2015GXNSFGA139003), the Bagui group of Guangxi province, and the project of State Key Laboratory for Chemistry and Molecular Engineering of Medicinal Resources, School of Chemistry and Pharmaceutical Science of Guangxi Normal University (CMEMR2014-A08).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics declarations
All procedures and experiments about human blood samples were performed in accordance with the Guidelines for Care and Use of Laboratory biological of Guangxi Normal University (Guilin, China) and approved by the biological Ethics Committee of China.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 3791 kb)
Rights and permissions
About this article
Cite this article
Li, Q., Liang, X., Mu, X. et al. Ratiometric fluorescent 3D DNA walker and catalyzed hairpin assembly for determination of microRNA. Microchim Acta 187, 365 (2020). https://doi.org/10.1007/s00604-020-04324-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-020-04324-5