Abstract
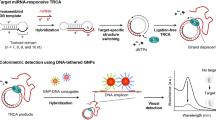
The authors describe a fluorometric switch assay for microRNA (miRNA). It is based on nonenzymatic ligation-rolling circle amplification (NELRCA). A click chemistry-mediated functionalized linear template was prepared by self-cycling. It is capable of forming a modified circular template (MLT) with a triazole linkage for amplified agent. MiRNA can recognize a binding region of the MLT, activates the NELRCA, and converts into rolling circle amplification products (RCPs). Subsequently, the added FAM-labeled signal probes (FAM-SPs) can hybridize to the RCPs. This RCPs/FAM-SPs duplex is less adsorbed onto the graphene oxide (GO) so that fluorescence (best measured at excitation/emission wavelengths of 490/520 nm) increases. In contrast to classical GO-based adsorption and displacement sensing system, the designed hybridization and adsorption fluorescent switch-on platform can reduce the assay time, and improve the detection efficiency of target. Under optimal conditions, the sensing platform has a detection limit as low as 0.75 fM. It can well discriminate target miRNA from other kinds of miRNA. Conceivably, this method can be extended to the trace detection of other nucleic acid biomarkers, and simultaneous recognition of multiple targets, thereby improving the early clinical diagnostic accuracy of cancer.

A hybridization and adsorption switch-on sensing system was fabricated on nonenzymatic ligation-rolling circle amplification. Modified circular template with 1,4-connect triazole linkage was adopted as an identified and amplified agent for high sensitive detection of microRNA.





Similar content being viewed by others
References
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Berezikov E, Cuppen E, Plasterk RH (2006) Approaches to microRNA discovery. Nat Genet 38(6s):S2
Válóczi A, Hornyik C, Varga N, Burgyán J, Kauppinen S, Havelda Z (2004) Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res 32(22):e175–e175
Pourmand N, Caramuta S, Villablanca A, Mori S, Karhanek M, Wang SX, Davis RW (2007) Branch migration displacement assay with automated heuristic analysis for discrete DNA length measurement using DNA microarrays. Proc Natl Acad Sci U S A 104(15):6146–6151
Du YQ, Yang XX, Li WL, Wang J, Huang CZ (2014) A cancer-targeted drug delivery system developed with gold nanoparticle mediated DNA–doxorubicin conjugates. RSC Adv 4(66):34830–34835
Liu H, Bei X, Xia Q, Fu Y, Zhang S, Liu M, Fan K, Zhang M, Yang Y (2016) Enzyme-free electrochemical detection of microRNA-21 using immobilized hairpin probes and a target-triggered hybridization chain reaction amplification strategy. Microchim Acta 183(1):297–304
Jia H, Li Z, Liu C, Cheng Y (2010) Ultrasensitive detection of microRNAs by exponential isothermal amplification. Angew Chem Int Ed 49(32):5498–5501
Du W, Lv M, Li J, Yu R, Jiang J (2016) A ligation-based loop-mediated isothermal amplification (ligation-LAMP) strategy for highly selective microRNA detection. Chem Commun 52(86):12721–12724
Zhou L, Wang J, Chen Z, Li J, Wang T, Zhang Z, Xie G (2017) A universal electrochemical biosensor for the highly sensitive determination of microRNAs based on isothermal target recycling amplification and a DNA signal transducer triggered reaction. Microchim Acta 184(5):1305–1313
Ali MM, Li F, Zhang Z, Zhang K, Kang D-K, Ankrum JA, Le XC, Zhao W (2014) Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem Soc Rev 43(10):3324–3341
Kim M-G, Park JY, Miao W, Lee J, Oh Y-K (2015) Polyaptamer DNA nanothread-anchored, reduced graphene oxide nanosheets for targeted delivery. Biomaterials 48:129–136
Li C, Qiu X, Hou Z, Deng K (2015) A dumbell probe-mediated rolling circle amplification strategy for highly sensitive transcription factor detection. Biosens Bioelectron 64:505–510
Zhuang J, Lai W, Chen G, Tang D (2014) A rolling circle amplification-based DNA machine for miRNA screening coupling catalytic hairpin assembly with DNAzyme formation. Chem Commun 50(22):2935–2938
Cheng Y, Zhang X, Li Z, Jiao X, Wang Y, Zhang Y (2009) Highly sensitive determination of microRNA using target-primed and branched rolling-circle amplification. Angew Chem Int Ed 121(18):3318–3322
Clausson C-M, Söderberg O, Arngården L, Ishaq O, Wählby C, Nilsson M, Krzywkowski T (2014) Compaction of rolling circle amplification products increases signal strength and integrity. Sci Rep 5:12317
Yi X, Li L, Peng Y, Guo L (2014) A universal electrochemical sensing system for small biomolecules using target-mediated sticky ends-based ligation-rolling circle amplification. Biosens Bioelectron 57:103–109
Presolski SI, Hong VP, Finn M (2011) Copper-Catalyzed Azide–Alkyne Click Chemistry for Bioconjugation. Curr Prot Chem Biol 3:153–162
Ishizuka T, Liu HS, Ito K, Xu Y (2016) Fluorescence imaging of chromosomal DNA using click chemistry. Sci Rep 6:33217
Qiu S, Li X, Xiong W, Xie L, Guo L, Lin Z, Qiu B, Chen G (2013) A novel fluorescent sensor for mutational p53 DNA sequence detection based on click chemistry. Biosens Bioelectron 41:403–408
El-Sagheer AH, Brown T (2014) Combined nucleobase and backbone modifications enhance DNA duplex stability and preserve biocompatibility. Chem Sci 5(1):253–259
Varizhuk AM, Kaluzhny DN, Novikov RA, Chizhov AO, Smirnov IP, Chuvilin AN, Tatarinova ON, Fisunov GY, Pozmogova GE, Florentiev VL (2013) Synthesis of triazole-linked oligonucleotides with high affinity to DNA complements and an analysis of their compatibility with biosystems. J Org Chem 78(12):5964–5969
Zhang X, Liu C, Sun L, Duan X, Li Z (2015) Lab on a single microbead: an ultrasensitive detection strategy enabling microRNA analysis at the single-molecule level. Chem Sci 6(11):6213–6218
Qiu X, Hildebrandt N (2015) Rapid and multiplexed microRNA diagnostic assay using quantum dot-based forster resonance energy transfer. ACS Nano 9(8):8449–8457
Yang C, Shi K, Dou B, Xiang Y, Chai Y, Yuan R (2015) In situ DNA-templated synthesis of silver nanoclusters for ultrasensitive and label-free electrochemical detection of microRNA. ACS Appl Mater Inte 7(2):1188–1193
Zhang Z, Wang Y, Zhang N, Zhang S (2016) Self-assembly of nucleic acid molecular aggregates catalyzed by a triple-helix probe for miRNA detection and single cell imaging. Chem Sci 7(7):4184–4189
Lu Z, Zhang L, Deng Y, Li S, He N (2012) Graphene oxide for rapid microRNA detection. Nano 4(19):5840–5842
Ryoo S-R, Lee J, Yeo J, Na H-K, Kim Y-K, Jang H, Lee JH, Han SW, Lee Y, Kim VN (2013) Quantitative and multiplexed microRNA sensing in living cells based on peptide nucleic acid and nano graphene oxide (PANGO). ACS Nano 7(7):5882–5891
Lu C, Huang P-JJ, Liu B, Ying Y, Liu J (2016) Comparison of graphene oxide and reduced graphene oxide for DNA adsorption and sensing. Langmuir 32(41):10776–10783
Cui L, Chen Z, Zhu Z, Lin X, Chen X, Yang CJ (2013) Stabilization of ssRNA on graphene oxide surface: an effective way to design highly robust RNA probes. Anal Chem 85(4):2269–2275
Park JS, Goo N-I, Kim D-E (2014) Mechanism of DNA adsorption and desorption on graphene oxide. Langmuir 30(42):12587–12595
Paul T, Bera SC, Agnihotri N, Mishra PP (2016) Single-molecule FRET studies of the hybridization mechanism during noncovalent adsorption and desorption of DNA on graphene oxide. J Phys Chem B 120(45):11628–11636
Zeng K, Li H, Peng Y (2017) Gold nanoparticle enhanced surface plasmon resonance imaging of microRNA-155 using a functional nucleic acid-based amplification machine. Microchim Acta 184(8):2637–2644
Borghei Y-S, Hosseini M, Ganjali MR (2017) Fluorescence based turn-on strategy for determination of microRNA-155 using DNA-templated copper nanoclusters. Microchim Acta 184(8):2671-2677
Yang J, Tang M, Diao W, Cheng W, Zhang Y, Yan Y (2016) Electrochemical strategy for ultrasensitive detection of microRNA based on MNAzyme-mediated rolling circle amplification on a gold electrode. Microchim Acta 183(11):3061–3067
Huang R, Liao Y, Zhou X, Xing D (2015) Toehold-mediated nonenzymatic amplification circuit on graphene oxide fluorescence switching platform for sensitive and homogeneous microRNA detection. Anal Chim Acta 888:162–172
Acknowledgements
This work was financially supported by the Natural Science Research Foundation of China (81672112, 81171415) and the Chongqing Yuzhong District Science and Technology Project (20140108).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author(s) declare that they have no competing interests.
Electronic supplementary material
ESM 1
(DOC 12644 kb)
Rights and permissions
About this article
Cite this article
Li, Y., Pu, Q., Li, J. et al. An “off-on” fluorescent switch assay for microRNA using nonenzymatic ligation-rolling circle amplification. Microchim Acta 184, 4323–4330 (2017). https://doi.org/10.1007/s00604-017-2475-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-017-2475-x