Abstract
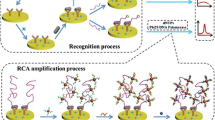
Okadaic acid (OA) is a low molecular weight marine toxin from shellfish which causes diarrheic shellfish poisoning (DSP). Due to its frequent occurrence, OA has become a serious threat to human health and seafood industry. The authors describe a competitive fluorophore-linked aptamer assay for OA that is based on rolling circle amplification (RCA). It consists of the following steps: (a) The wells of a microplate are modified by fixing the OA aptamer on their surface; (b) The aptamer is hybridized with an aptamer-complementary sequence-primer complex; (c) the RCA reaction is performed; (d) the FAM labelled signal probe is added. OA competes with the detection probe for the immobilized aptamer. After the competitive reaction has occurred, the supernatants containing released detection probes are removed and then read with a microplate reader. This method, unlike in competitive assays where the signals negatively correlate with OA concentrations, has a positive correlation between fluorescence intensity and OA concentration. The optimized assay has a lower detection limit (1 pg·mL−1) and a wider linear range (from 1 pg·mL−1 to 100 ng·mL−1) owning to signal amplification via RCA. It also is highly specific, repeatable, has good recovery and can be used to detect OA in seafood.

Schematic of a microplate assay for okadaic acid assisted by rolling circle amplification (RCA) and using a fluorophore-linked aptamer. The signal intensities are directly proportional to the concentrations of OA. The assay without RCA was also performed and compared to.



Similar content being viewed by others
References
Tachibana K, Scheuer PJ, Tsukitani Y, Kikuchi H, Van Engen D, Clardy J, Gopichand Y, Schmitz FJ (1981) Okadaic acid, a cytotoxic polyether from two marine sponges of the genus Halichondria. J Am Chem Soc 103(9):2469–2471. doi:10.1021/ja00399a082
Campàs M, Prieto-Simón B, Marty J-L (2007) Biosensors to detect marine toxins: assessing seafood safety. Talanta 72(3):884–895. doi:10.1016/j.talanta.2006.12.036
Paredes I, Rietjens IMCM, Vieites JM, Cabado AG (2011) Update of risk assessments of main marine biotoxins in the European Union. Toxicon 58(4):336–354. doi:10.1016/j.toxicon.2011.07.001
Fujiki H, Suganuma M (1993) Tumor promotion by inhibitors of ProteinZ phosphatases 1 and 2A: the Okadaic acid class of compounds. Adv Cancer Res 61:143–194. doi:10.1016/s0065-230x(08)60958-6
Meštrović V, Pavela-Vrančič M (2003) Inhibition of alkaline phosphatase activity by okadaic acid, a protein phosphatase inhibitor. Biochimie 85(7):647–650. doi:10.1016/s0300-9084(03)00135-4
Valdiglesias V, Prego-Faraldo M, Pásaro E, Méndez J, Laffon B (2013) Okadaic acid: more than a diarrheic toxin. Mar Drugs 11(11):4328–4349. doi:10.3390/md11114328
Kamat PK, Rai S, Swarnkar S, Shukla R, Nath C (2014) Molecular and cellular mechanism of Okadaic acid (OKA)-induced neurotoxicity: a novel tool for Alzheimer’s disease therapeutic application. Mol Neurobiol 50(3):852–865. doi:10.1007/s12035-014-8699-4
Deeds JR, Wiles K, Heideman GB, White KD, Abraham A (2010) First U.S. report of shellfish harvesting closures due to confirmed okadaic acid in Texas gulf coast oysters. Toxicon 55(6):1138–1146. doi:10.1016/j.toxicon.2010.01.003
Stabell O, Steffenak I, Aune T (1992) An evaluation of the mouse bioassay applied to extracts of ‘diarrhoetic’shellfish toxins. Food Chem Toxicol 30(2):139–144. doi:10.1016/0278-6915(92)90149-F
Cañete E, Diogène J (2008) Comparative study of the use of neuroblastoma cells (neuro-2a) and neuroblastoma×glioma hybrid cells (NG108-15) for the toxic effect quantification of marine toxins. Toxicon 52(4):541–550. doi:10.1016/j.toxicon.2008.06.028
Nogueiras MJ, Gago-Martínez A, Paniello AI, Twohig M, James KJ, Lawrence JF (2003) Comparison of different fluorimetric HPLC methods for analysis of acidic polyether toxins in marine phytoplankton. Anal Bioanal Chem 377(7–8):1202–1206. doi:10.1007/s00216-003-2221-6
Wu Z, Wang B, Sun Y, Liu Y (2015) Improvement of determination method of okadaic acid in shellfish by liquid chromatography-tandem mass spectrometry. Journal of Food Safety and Quality 6(1):265–271
Liu B-H, Hung C-T, Lu C-C, Chou H-N, Yu F-Y (2014) Production of monoclonal antibody for Okadaic acid and its utilization in an ultrasensitive enzyme-linked immunosorbent assay and one-step Immunochromatographic strip. J Agric Food Chem 62(6):1254–1260. doi:10.1021/jf404827s
Sassolas A, Catanante G, Hayat A, Marty J-L (2011) Development of an efficient protein phosphatase-based colorimetric test for okadaic acid detection. Anal Chim Acta 702(2):262–268. doi:10.1016/j.aca.2011.07.002
Bunka DHJ, Stockley PG (2006) Aptamers come of age – at last. Nat Rev Microbiol 4(8):588–596. doi:10.1038/nrmicro1458
Gotrik MR, Feagin TA, Csordas AT, Nakamoto MA, Soh HT (2016) Advancements in aptamer discovery technologies. Acc Chem Res 49(9):1903–1910. doi:10.1021/acs.accounts.6b00283
Yuan J, Wu S, Duan N, Ma X, Xia Y, Chen J, Ding Z, Wang Z (2014) A sensitive gold nanoparticle-based colorimetric aptasensor for Staphylococcus aureus. Talanta 127:163–168. doi:10.1016/j.talanta.2014.04.013
Huang Y, Zhang H, Chen X, Wang X, Duan N, Wu S, Xu B, Wang Z (2015) A multicolor time-resolved fluorescence aptasensor for the simultaneous detection of multiplex Staphylococcus aureus enterotoxins in the milk. Biosens Bioelectron 74:170–176. doi:10.1016/j.bios.2015.06.046
Hao L, Duan N, Wu S, Xu B, Wang Z (2015) Chemiluminescent aptasensor for chloramphenicol based on N-(4-aminobutyl)-N-ethylisoluminol-functionalized flower-like gold nanostructures and magnetic nanoparticles. Anal Bioanal Chem 407(26):7907–7915. doi:10.1007/s00216-015-8957-y
Li A, Tang L, Song D, Song S, Ma W, Xu L, Kuang H, Wu X, Liu L, Chen X, Xu C (2016) A SERS-active sensor based on heterogeneous gold nanostar core–silver nanoparticle satellite assemblies for ultrasensitive detection of aflatoxinB1. Nano 8(4):1873–1878. doi:10.1039/c5nr08372a
Mishra RK, Hayat A, Catanante G, Ocaña C, Marty J-L (2015) A label free aptasensor for Ochratoxin a detection in cocoa beans: an application to chocolate industries. Anal Chim Acta 889:106–112. doi:10.1016/j.aca.2015.06.052
Gu H, Duan N, Wu S, Hao L, Xia Y, Ma X, Wang Z (2016) Graphene oxide-assisted non-immobilized SELEX of okdaic acid aptamer and the analytical application of aptasensor. Sci Rep 6:21665. doi:10.1038/srep21665
Wang RE, Zhang Y, Cai J, Cai W, Gao T (2011) Aptamer-based fluorescent biosensors. Curr Med Chem 18(27):4175–4184. doi:10.2174/092986711797189637
Li F, Zhang H, Wang Z, Newbigging AM, Reid MS, Li X-F, Le XC (2015) Aptamers facilitating amplified detection of biomolecules. Anal Chem 87(1):274–292. doi:10.1021/ac5037236
Fire A, Xu S-Q (1995) Rolling replication of short DNA circles. Proc Nat Acad Sci USA 92(10):4641–4645. doi:10.1073/pnas.92.10.4641
Shen B, Li J, Cheng W, Yan Y, Tang R, Li Y, Ju H, Ding S (2014) Electrochemical aptasensor for highly sensitive determination of cocaine using a supramolecular aptamer and rolling circle amplification. Microchim Acta 182(1–2):361–367. doi:10.1007/s00604-014-1333-3
Yang J, Tang M, Diao W, Cheng W, Zhang Y, Yan Y (2016) Electrochemical strategy for ultrasensitive detection of microRNA based on MNAzyme-mediated rolling circle amplification on a gold electrode. Microchim Acta 183(11):3061–3067. doi:10.1007/s00604-016-1958-5
Zou L, Wang Q, Tong M, Li H, Wang J, Hu N, Wang P (2016) Detection of diarrhetic shellfish poisoning toxins using high-sensitivity human cancer cell-based impedance biosensor. Sensors Actuators B Chem 222:205–212. doi:10.1016/j.snb.2015.08.061
Garibo D, Devic E, Marty J-L, Diogène J, Unzueta I, Blázquez M, Campàs M (2012) Conjugation of genetically engineered protein phosphatases to magnetic particles for okadaic acid detection. J Biotechnol 157(1):89–95. doi:10.1016/j.jbiotec.2011.11.020
Zhou J, Qiu X, Su K, Xu G, Wang P (2016) Disposable poly (o-aminophenol)-carbon nanotubes modified screen print electrode-based enzyme sensor for electrochemical detection of marine toxin okadaic acid. Sensors Actuators B Chem 235:170–178. doi:10.1016/j.snb.2016.05.067
Sassolas A, Catanante G, Hayat A, Stewart LD, Elliott CT, Marty JL (2013) Improvement of the efficiency and simplification of ELISA tests for rapid and ultrasensitive detection of okadaic acid in shellfish. Food Control 30(1):144–149. doi:10.1016/j.foodcont.2012.05.028
Vdovenko MM, Hung C-T, Sakharov IY, Yu F-Y (2013) Determination of okadaic acid in shellfish by using a novel chemiluminescent enzyme-linked immunosorbent assay method. Talanta 116:343–346. doi:10.1016/j.talanta.2013.05.057
Garibo D, Campbell K, Casanova A, de la Iglesia P, Fernández-Tejedor M, Diogène J, Elliott CT, Campàs M (2014) SPR immunosensor for the detection of okadaic acid in mussels using magnetic particles as antibody carriers. Sensors Actuators B Chem 190:822–828. doi:10.1016/j.snb.2013.09.037
Eissa S, Ng A, Siaj M, Tavares AC, Zourob M (2013) Selection and identification of DNA aptamers against Okadaic acid for Biosensing application. Anal Chem 85(24):11794–11801. doi:10.1021/ac402220k
Acknowledgements
This work was partly supported by the National Science and Technology Support Program of China (2015BAD17B02), the Key Research and Development Program of Jiangsu Province (BE2016306), the National Natural Science Foundation of China (31401576, 31401575), the Technology R&D Program of Suzhou (SYN201513), the National Grain Special Public-Funded Program of China (201513006), and Synergetic Innovation Center of Food Safety and Quality control of Jiangsu Province.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplementary Material
(PDF 1.36 mb)
Rights and permissions
About this article
Cite this article
Gu, H., Hao, L., Duan, N. et al. A competitive fluorescent aptasensor for okadaic acid detection assisted by rolling circle amplification. Microchim Acta 184, 2893–2899 (2017). https://doi.org/10.1007/s00604-017-2293-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-017-2293-1