Abstract
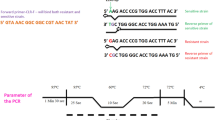
Helicobacter pylori infection can be eradicated in up to 90 % of patients using current combination of triple therapies, of which clarithromycin is a key component. The development of clarithromycin resistance in H. pylori is recognized as a significant contributing factor in treatment failure. The aim of this study is to compare two fast and direct diagnostic methods to evaluate clarithromycin resistance in gastric biopsy specimens. This cross-sectional descriptive study was performed on 150 gastric biopsy specimens. Analysis for clarithromycin resistance was performed by real-time polymerase chain reaction (PCR) assay for A2144G, A2143G, A2143C, and A2142G point mutations by specific probes. Also, A2142G and A2143G point mutations were detected by PCR-RFLP method and using BsaI and MboII restriction enzymes. Out of 150 samples, 96 (64 %) clarithromycin-sensitive- and 54 (36 %) clarithromycin-resistant strains were detected by TaqMan real-time PCR assay. Thirty-seven (24.67 %) resistant strains were detected by PCR-RFLP method. Results showed that real-time PCR assay has enough accuracy to identify clarithromycin resistance and their mutations in a short time.

Similar content being viewed by others
References
Alfaresi MS, Abdulsalam AI, Elkoush AA (2007) Analysis of Helicobacter pylori antimicrobial susceptibility and virulence genes in gastric mucosal biopsies in the United Arab Emirates. Indian J Gastroenterol 26:221–224
Ben Mansour K, Burucoa C, Zribi M, Masmoudi A, Karoui S, Kallel L, Chouaib S, Matri S, Fekih M, Zarrouk S, Labbene M, Boubaker J, Cheikh I, Hriz MB, Siala N, Ayadi A, Filali A, Mami NB, Najjar T, Maherzi A, Sfar MT, Fendri C (2010) Primary resistance to clarithromycin, metronidazole and amoxicillin of Helicobacter pylori isolated from Tunisian patients with peptic ulcers and gastritis: a prospective multicentre study. Ann Clin Microbiol and Antimicrob 9:22. Available online at: http://www.ann-clinmicrob.com/content/9/1/22
Burucoa C, Garnier M, Silvain C, Fauchere JL (2008) Quadruplex real-time PCR assay using allele-specific scorpion primers for detection of mutations conferring clarithromycin resistance to Helicobacter pylori. J Clin Microbiol 46:2320–2326
Chisholm SA, Owen RJ, Teare EL, Saverymuttu S (2001) PCR-based diagnosis of Helicobacter pylori infection and real-time determination of clarithromycin resistance directly from human gastric biopsy samples. J Clin Microbiol 39:1217–1220
Cirak MY, Akyön Y, Mégraud F (2007) Diagnosis of Helicobacter pylori. Helicobacter 12:4–9
De Francesco V, Margiotta M, Zullo A, Hassan C, Valle ND, Burattini O, Cea U, Stoppino G, Amoruso A, Stella F, Morini S, Panella C, Ierardi E (2006) Primary clarithromycin resistance in Italy assessed on Helicobacter pylori DNA sequences by TaqMan real-time polymerase chain reaction. Aliment Pharmacol Ther 23:429–435
De Francesco V, Zullo A, Ierardi E, Giorgio F, Perna F, Hassan C, Morini S, Panella C, Vaira D (2010) Phenotypic and genotypic Helicobacter pylori clarithromycin resistance and therapeutic outcome: benefits and limits. J Antimicrob Chemother 65:327–332
Demir M, Gokturk HS, Ozturk NA, Arslan H, Serin E, Yilmaz U (2009) Clarithromycin resistance and efficacy of clarithromycin-containing triple eradication therapy for Helicobacter pylori infection in type 2 diabetes mellitus patients. South Med J 102:116–120
Elviss NC, Lawson AJ, Owen RJ (2004) Application of 3′-mismatched reverse primer PCR compared with real-time PCR and PCR-RFLP for the rapid detection of 23S rDNA mutations associated with clarithromycin resistance in Helicobacter pylori. Int J Antimicrob Agents 23:349–355
Falsafi T, Mobasheri F, Nariman F, Najafi M (2004) Susceptibilities to different antibiotics of Helicobacter pylori strains isolated from patients at the Pediatric Medical Center of Tehran, Iran. J Clin Microbiol 42:387–389
Kargar M, Baghernejad M, Doosti A (2010) Role of NADPH-insensitive nitroreductase gene to metronidazole resistance of Helicobacter pylori strains. DARU 18:137–140
Kargar M, Souod N, Ghorbani-Dalini S, Doosti A, Rezaeian AA (2011a) Evaluation of cagA tyrosine phosphorylation DNA motifs in Helicobacter pylori isolates from gastric disorder patients in west of Iran. Sci Res Essays 6:6454–6458
Kargar M, Souod N, Doosti A, Ghorbani-Dalini S (2011b) Prevalence of Helicobacter pylori vacuolating cytotoxin A gene as a predictive marker for different gastrodoudenal diseases. Iran J Clin Infect Dis 6:85–89
Kargar M, Baghernejad M, Doosti S, Ghorbani-Dalini S (2011c) Clarithromycin resistance and 23S rRNA mutations in Helicobacter pylori isolates in Iran. Afr J Microbiol Res 5:853–856
Kargar M, Ghorbani-Dalini S, Doosti A, Baghernejad M (2011d) Molecular assessment of clarithromycin resistant Helicobacter pylori strains using rapid and accurate PCR-RFLP method in gastric specimens in Iran. Afr J Biotechnol 10:7675–7678
Kargar M, Ghorbani-Dalini S, Doosti A, Souod N (2012) Real-time PCR for Helicobacter pylori quantification and detection of clarithromycin resistance in gastric tissues from patients with gastrointestinal disorders. Res Microbiol 163:109–113
Kato S, Fujimura S, Udagawa H, Shimizu T, Maisawa S, Ozawa K, Linuma K (2002) Antibiotic resistance of Helicobacter pylori strains in Japanese children. J Clin Microbiol 40:649–653
Lascols C, Lamarque D, Costa JM, Copie-Bergman C, Glaunec JML, Deforges L, Soussy CJ, Petit JC, Delchier JC, Tankovic J (2003) Fast and accurate quantitative detection of Helicobacter pylori and identification of clarithromycin resistance mutations in H. pylori isolates from gastric biopsy specimens by real-time PCR. J Clin Microbiol 41:4573–4577
Liu Z, Shen J, Zhang L, Shen L, Li Q, Zhang B, Zhou J, Gu L, Feng G, Ma J, You WC, Deng D (2008) Prevalence of A2143G mutation of H. pylori-23S rRNA in Chinese subjects with and without clarithromycin use history. BMC Microbiol 8:81. doi:10.1186/1471-2180-8-81
Lottspeich C, Schwarzer A, Panthel K, Koletzko S, Russmann H (2007) Evaluation of the novel Helicobacter pylori ClariRes real-time PCR assay for detection and clarithromycin susceptibility testing of H. pylori in stool specimens from symptomatic children. J Clin Microbiol 45:1718–1722
Marais A, Monteiro L, Occhialini A, Pina M, Lamouliatte H, Mégraud F (1999) Direct detection of Helicobacter pylori resistance to macrolides by a polymerase chain reaction/DNA enzyme immunoassay in gastric biopsy specimens. Gut 44:463–467
Matsuoka M, Yoshida Y, Hayakawa K, Fukuchi S, Sugano K (1999) Simultaneous colonisation of Helicobacter pylori with and without mutations in the 23S rRNA gene in patients with no history of clarithromycin exposure. Gut 45:503–507
Oleastro M, Menard A, Santos A, Lamouliatte H, Monteiro L, Barthelemy P, Mégraud F (2003) Real-time PCR assay for rapid and accurate detection of point mutations conferring resistance to clarithromycin in Helicobacter pylori. J Clin Microbiol 41:397–402
Pan ZJ, Su WW, Tytgat GNJ, Dankert J, Van Der Ende AA (2002) Assessment of clarithromycin-resistant Helicobacter pylori among patients in Shanghai and Guangzhou, China, by Primer-Mismatch PCR. J Clin Microbiol 40:259–261
Pina M, Occhialini A, Monteiro L, Doermann HP, Megraud F (1998) Detection of point mutations associated with resistance of Helicobacter pylori to clarithromycin by hybridization in liquid phase. J Clin Microbiol 36:3285–3290
Razaghi M, Boutorabi SM, Mirjalili A, Norolahi S, Hashemi M, Jalalian M (2010) Diagnosis of Helicobacter pylori infection by ELISA stool antigen and comparison with the other diagnostic methods. Health Med 4:545–551
Ribeiro ML, Ecclissato CC, Mattos RG, Mendonca S, Pedrazzoli J Jr (2007) Quantitative real-time PCR for the clinical detection of Helicobacter pylori. Genet Mol Biol 30:431–434
Schabereiter-Gurtner C, Hirschl AM, Dragosics B, Hufnagl P, Puz S, Kovach Z, Rotter M, Makristathis A (2004) Novel real-time PCR assay for detection of Helicobacter pylori infection and simultaneous clarithromycin susceptibility testing of stool and biopsy specimens. J Clin Microbiol 42:4512–4518
Taneike I, Suzuki K, Nakagawa S, Yamamoto T (2004) Intrafamilial spread of the same clarithromycin-resistant Helicobacter pylori infection confirmed by molecular analysis. J Clin Microbiol 42:3901–3903
Taylor DE, Ge Z, Purych D, Lo T, Hiratsuka K (1997) Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob Agents Chemother 41:2621–2628
Van Der Ende A, Van Doorn LJ, Rooijakkers S, Feller M, Tytgat GNJ, Dankert J (2001) Clarithromycin-susceptible and -resistant Helicobacter pylori isolates with identical randomly amplified polymorphic DNA-PCR genotypes cultured from single gastric biopsy specimens prior to antibiotic therapy. J Clin Microbiol 39:2648–2651
Woo HY, Park DI, Park H, Kim MK, Kim DH, Kim IS, Kim YJ (2009) Dual-priming oligonucleotide-based multiplex PCR for the detection of Helicobacter pylori and determination of clarithromycin resistance with gastric biopsy specimens. Helicobacter 14:22–28
Yilmaz Ö, Demiray E (2007) Clinical role and importance of fluorescence in situ hybridization method in diagnosis of H. pylori infection and determination of clarithromycin resistance in H pylori eradication therapy. World J Gastroenterol 13:671–675
Acknowledgments
The authors offer their thanks and appreciation to the Islamic Azad University, Jahrom and Shahrekord branches for their executive protection of this project.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kargar, M., Doosti, A. & Ghorbani-Dalini, S. Detection of four clarithromycin resistance point mutations in Helicobacter pylori: comparison of real-time PCR and PCR-RFLP methods. Comp Clin Pathol 22, 1007–1013 (2013). https://doi.org/10.1007/s00580-012-1519-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00580-012-1519-1