Abstract
Transcriptomics and genomics data recently obtained from the arbuscular mycorrhizal (AM) fungus Rhizophagus irregularis have offered new opportunities to decipher the contribution of the fungal partner to the establishment of the symbiotic association. The large number of genes which do not show similarity to known proteins witnesses the uniqueness of this group of plant-associated fungi. In this work, we characterize a gene that was called RiPEIP1 (Preferentially Expressed In Planta). Its expression is strongly induced in the intraradical phase, including arbuscules, and follows the expression profile of the Medicago truncatula phosphate transporter MtPT4, a molecular marker of a functional symbiosis. Indeed, mtpt4 mutant plants, which exhibit low mycorrhizal colonization and an accelerated arbuscule turnover, also show a reduced RiPEIP1 mRNA abundance. To further characterize RiPEIP1, in the absence of genetic transformation protocols for AM fungi, we took advantage of two different fungal heterologous systems. When expressed as a GFP fusion in yeast cells, RiPEIP1 localizes in the endomembrane system, in particular to the endoplasmic reticulum, which is consistent with the in silico prediction of four transmembrane domains. We then generated RiPEIP1-expressing strains of the fungus Oidiodendron maius, ericoid endomycorrhizal fungus for which transformation protocols are available. Roots of Vaccinium myrtillus colonized by RiPEIP1-expressing transgenic strains showed a higher mycorrhization level compared to roots colonized by the O. maius wild-type strain, suggesting that RiPEIP1 may regulate the root colonization process.







Similar content being viewed by others
References
Abbà S, Khouja HR, Martino E, Archer DB, Perotto S (2009) SOD1-targeted gene disruption in the ericoid mycorrhizal fungus Oidiodendron maius reduces conidiation and the capacity for mycorrhization. Mol Plant Microbe Interact 22:1412–1421. doi:10.1094/MPMI-22-11-1412
Allen JW, Shachar-Hill Y (2009) Sulfur transfer through an arbuscular mycorrhiza. Plant Physiol 149:549–560. doi:10.1104/pp.108.129866
Balestrini R, Gómez-Ariza J, Lanfranco L, Bonfante P (2007) Laser microdissection reveals that transcripts for five plant and one fungal phosphate transporter genes are contemporaneously present in arbusculated cells. Mol Plant Microbe Interact 20:1055–1062. doi:10.1094/MPMI-20-9-1055
Bécard G, Fortin JA (1988) Early events of vesicular–arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol 108:211–218. doi:10.1111/j.1469-8137.1988.tb03698.x
Belmondo S, Fiorilli V, Pérez-Tienda J, Ferrol N, Marmeisse R, Lanfranco L (2014) A dipeptide transporter from the arbuscular mycorrhizal fungus Rhizophagus irregularis is upregulated in the intraradical phase. Front Plant Sci 3(5):436. doi:10.3389/fpls.2014.00436
Blaudez D, Kohler A, Martin F, Sanders D, Chalot M (2003) Poplar metal tolerance protein 1 confers zinc tolerance and is an oligomeric vacuolar zinc transporter with an essential leucine zipper motif. Plant Cell 1512:2911–28. doi:10.1105/tpc.017541
Bonfante P, Genre A (2015) Arbuscular mycorrhizal dialogues: do you speak 'plantish' or 'fungish'? Trends Plant Sci 20:150–154. doi:10.1016/j.tplants.2014.12.002
Bonfante P, Requena N (2011) Dating in the dark: how roots respond to fungal signals to establish arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol 14:451–457. doi:10.1016/j.pbi.2011.03.014
Breuninger M, Requena N (2004) Recognition events in AM symbiosis: analysis of fungal gene expression at the early appressorium stage. Fungal Genet Biol 41:794–804. doi:10.1016/j.fgb.2004.04.002
Cappellazzo G, Lanfranco L, Bonfante P (2007) A limiting source of organic nitrogen induces specific transcriptional responses in the extraradical structures of the endomycorrhizal fungus Glomus intraradices. Curr Genet 51:59–70. doi:10.1007/s00294-006-0101-2
Ehinger MO, Croll D, Koch AM, Sanders IR (2012) Significant genetic and phenotypic changes arising from clonal growth of a single spore of an arbuscular mycorrhizal fungus over multiple generations. New Phytol 196:853–561. doi:10.1111/j.1469-8137.2012.04278.x
Fiorilli V, Lanfranco L, Bonfante P (2013) The expression of GintPT, the phosphate transporter of Rhizophagus irregularis, depends on the symbiotic status and phosphate availability. Planta 237:1267–1277. doi:10.1007/s00425-013-1842-z
Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG (2005) Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17:3489–3499. doi:10.1105/tpc.105.035410
Genre A, Chabaud M, Faccio A, Barker DG, Bonfante P (2008) Pre-penetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20:1407–1420. doi:10.1105/tpc.108.059014
Gianinazzi S, Gollotte A, Binet MN, van Tuinen D, Redecker D, Wipf D (2010) Agroecology: the key role of arbuscular mycorrhizas in ecosystem services. Mycorrhiza 20:519–530. doi:10.1007/s00572-010-0333-3
Gietz D, St Jean A, Woods RA, Schies RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20:1425
Giovannetti M, Sbrana C, Avio L, Citernesi AS, Logi C (1993) Differential hyphal morphogenesis in arbuscular mycorrhizal fungi during pre-infection stages. New Phytol 125:587–593. doi:10.1111/j.1469-8137.1993.tb03907.x
González-Guerrero M, Oger E, Benabdellah K, Azcón-Aguilar C, Lanfranco L, Ferrol N (2010) Characterization of a CuZn superoxide dismutase gene in the arbuscular mycorrhizal fungus Glomus intraradices. Curr Genet 56:265–74. doi:10.1007/s00294-010-0298-y
Govindarajulu M, Pfeffer PE, Jin HR, Abubaker J, Douds DD, Allen JW, Bücking H, Lammers PJ, Shachar-Hil Y (2005) Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 435:819–823. doi:10.1038/nature03610
Gutjahr C, Parniske M (2013) Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu Rev Cell Dev Biol 29:593–617. doi:10.1146/annurev-cellbio-101512-122413
Halary S, Malik SB, Lildhar L, Slamovits CH, Hijri M, Corradi N (2011) Conserved meiotic machinery in Glomus spp., a putatively ancient asexual fungal lineage. Genome Biol Evol 3:950–958. doi:10.1093/gbe/evr089
Halary S, Daubois L, Terrat Y, Ellenberger S, Wöstemeyer J, Hijri M (2013) Mating type gene homologues and putative sex pheromone-sensing pathway in arbuscular mycorrhizal fungi, a presumably asexual plant root symbiont. Plos One 8(11), e80729. doi:10.1371/journal.pone.0080729
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:9
Harrison MJ, Dewbre GR, Liu J (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14:2413–2429. doi:10.1105/tpc.004861
Helber N, Requena N (2008) Expression of the fluorescence markers DsRed and GFP fused to a nuclear localization signal in the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol 177:537–548. doi:10.1111/j.1469-8137.2007.02257.x
Helber N, Wippel N, Schaarschmidt S, Hause B, Requena N (2011) A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp. is crucial for the symbiotic relationship with plants. Plant Cell 23:3812–3823. doi:10.1105/tpc.111.089813
Hijri M, Sanders IR (2005) Low gene copy number shows that arbuscular mycorrhizal fungi inherit genetically different nuclei. Nature 433:160–163. doi:10.1038/nature03069
Hohnjec N, Vieweg MF, Pühler A, Becker A, Küster H (2005) Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol 137:1283–1301. doi: 10.1104/pp.104.056572
Jackson MR, Nilsson T, Peterson PA (1990) Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J 9:3153–3162
Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ (2007) A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci U S A 104:1720–1725. doi:10.1073/pnas.0608136104
Javot H, Penmetsa RV, Breuillin F, Bhattarai KK, Noar RD, Gomez SK, Zhang Q, Cook DR, Harrison MJ (2011) Medicago truncatula mtpt4 mutants reveal a role for nitrogen in the regulation of arbuscule degeneration in arbuscular mycorrhizal symbiosis. Plant J 68:954–965. doi:10.1111/j.1365-313X.2011.04746.x
Kikuchi Y, Hijikata N, Yokoyama K, Ohtomo RY, Handa Y, Kawaguchi M, Katsuharu Saito K, Ezawa T (2014) Polyphosphate accumulation is driven by transcriptome alterations that lead ners-synchronous and near-equivalent uptake of inorganic cations in an arbuscular mycorrhizal fungus. New Phytol 204:638–649. doi:10.1111/nph.12937
Kloppholz S, Kuhn H, Requena N (2011) A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol 21:1204–1209. doi:10.1016/j.cub.2011.06.044
Kobae Y, Hata S (2010) Dynamics of periarbuscular membranes visualized with a fluorescent phosphate transporter in arbuscular mycorrhizal roots of rice. Plant Cell Physiol 51:341–53. doi:10.1093/pcp/pcq013
Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, Canbäck B, Choi C, Cichocki N, Clum A, Colpaert J, Copeland A, Costa MD, Doré J, Floudas D, Gay G, Girlanda M, Henrissat B, Herrmann S, Hess J, Högberg N, Johansson T, Khouja HR, LaButti K, Lahrmann U, Levasseur A, Lindquist EA, Lipzen A, Marmeisse R, Martino E, Murat C, Ngan CY, Nehls U, Plett JM, Pringle A, Ohm RA, Perotto S, Peter M, Riley R, Rineau F, Ruytinx J, Salamov A, Shah F, Sun H, Tarkka M, Tritt A, Veneault-Fourrey C, Zuccaro A, Tunlid A, Grigoriev A IV, Hibbett DS, Martin F (2015) Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet 47:410–415
Kormanik PP, McGraw AC (1982) Quantification of vesicular-arbuscular mycorrhizae in plant roots. In: Schenk NC (ed) Methods and principles of mycorrhizal research American Phytopathological Society. St Paul, MN, pp p37–47
Lanfranco L, Young JP (2012) Genetic and genomic glimpses of the elusive arbuscular mycorrhizal fungi. Curr Opin Plant Biol 15:454–461. doi:10.1016/j.pbi.2012.04.003
Li T, Hu YJ, Hao ZP, Li H, Wang YS, Chen BD (2013) First cloning and characterization of two functional aquaporin genes from an arbuscular mycorrhizal fungus Glomus intraradices. New Phytol 197:617–30. doi:10.1111/nph.12011
Lin K, Limpens E, Zhang Z, Ivanov S, Saunders DG, Mu D, Pang E, Cao H, Cha H, Lin T, Zhou Q, Shang Y, Li Y, Sharma T, van Velzen R, de Ruijter N, Aanen DK, Win J, Kamoun S, Bisseling T, Geurts R, Huang S (2014) Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet 10, e1004078. doi:10.1371/journal.pgen.1004078
Martin F, Kohler A, Murat C, Balestrini R, Coutinho PM, Jaillon O, Montanini B, Morin E, Noel B, Percudani R, Porcel B, Rubini A, Amicucci A, Amselem J, Anthouard V, Arcioni S, Artiguenave F, Aury JM, Ballario P, Bolchi A, Brenna A, Brun A, Buée M, Cantarel B, Chevalier G, Couloux A, Da Silva C, Denoeud F, Duplessis S, Ghignone S, Hilselberger B, Iotti M, Marçais B, Mello A, Miranda M, Pacioni G, Quesneville H, Riccioni C, Ruotolo R, Splivallo R, Stocchi V, Tisserant E, Viscomi AR, Zambonelli A, Zampieri E, Henrissat B, Lebrun MH, Paolocci F, Bonfante P, Ottonello S, Wincker P (2010) Périgord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 464:1033–1038. doi:10.1038/nature08867
Martino E, Turnau K, Girlanda M, Bonfante P, Perotto S (2000) Ericoid mycorrhizal fungi from heavy metal polluted soils: their identification and growth in the presence of zinc ions. Mycol Res 104:338–344. doi:10.1017/S0953756299001252
Martino E, Murat C, Vallino M, Bena A, Perotto S, Spanu P (2007) Imaging mycorrhizal fungal transformants that express EGFP during ericoid endosymbiosis. Curr Genet 52:65–75. doi:10.1007/s00294-007-0139-9
Pawlowska TE, Taylor JW (2004) Organization of genetic variation in individuals of arbuscular mycorrhizal fungi. Nature 427:733–737. doi:10.1038/nature02290
Pérez-Tienda J, Balestrini R, Fiorilli V, Azcón-Aguilar C, Ferrol N (2011) GintAMT2, a new member of the ammonium transporter family in the arbuscular mycorrhizal fungus Glomus intraradices. Fungal Genet Biol 48:1044–1055. doi:10.1016/j.fgb.2011.08.003
Pfeffer PE, Douds DD, Bécard G, Shachar-Hill Y (1999) Carbon uptake and the metabolism and transport of lipids in an arbuscular mycorrhiza. Plant Physiol 120:587–598. doi:10.1104/pp.120.2.587
Pumplin N, Harrison MJ (2009) Live-cell imaging reveals periarbuscular membrane domains and organelle location in Medicago truncatula roots during arbuscular mycorrhizal symbiosis. Plant Physiol 151:809–819. doi:10.1104/pp.109.141879
Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel CA (1987) Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56:117–24
Redecker D, Schüssler A, Stockinger H, Stürmer SL, Morton JB, Walker C (2013) An evidence-based consensus for the classification of arbuscular mycorrhizal fungi. (Glomeromycota) Mycorrhiza 23:515–531. doi:10.1007/s00572-013-0486-y
Requena N, Mann P, Hampp R, Franken P (2002) Early developmentally regulated genes in the arbuscular mycorrhizal fungus Glomus mosseae: GmGIN1, a novel gene encoding a protein with homology to the C-terminus of metazoan hedgehog proteins. Plant Soil 244:129–139
Riley R, Salamov AA, Brown DW, Nagy LG, Floudas D, Held BW, Levasseur A, Lombard V, Morin E, Otillar R, Lindquist EA, Sun H, LaButti KM, Schmutz J, Jabbour D, Luo H, Baker SE, Pisabarro AG, Walton JD, Blanchette RA, Henrissat B, Martin F, Cullen D, Hibbett DS, Grigoriev IV (2014) Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc Natl Acad Sci U S A 111:9923–9928. doi:10.1073/pnas.1400592111
Ruzicka D, Chamala S, Barrios-Masias FH, Martin F, Smith S, Jackson LE, Brad Barbazuk W, Schachtman DP (2013) Inside arbuscular mycorrhizal roots—molecular probes to understand the symbiosis. Plant genome 6:1–13. doi:10.3835/plantgenome2012.06.0007
Salvioli A, Bonfante P (2013) Systems biology and "omics" tools: a cooperation for next-generation mycorrhizal studies. Plant Sci 204:107–114. doi:10.1016/j.plantsci.2013.01.001
Salvioli A, Ghignone S, Novero M, Navazio L, Venice F, Bagnaresi P, Bonfante P (2016) Symbiosis with an endobacterium increases the fitness of a mycorrhizal fungus, raising its bioenergetic potential. ISME J 10:130–144. doi:10.1038/ismej201591
Sambrook J, Russel DW (2001) Molecular cloning: a laboratory manual, volume 1, 2, 3 Cold Spring Harbour Laboratory Press. Cold Spring Harbour, New York
Schüßler A, Schwarzott D, Walker C (2001) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 105:1413–1421
Spanu PD, Abbott JC, Amselem J, Burgis TA, Soanes DM, Stüber K, van Ver Loren, Themaat E, Brown JK, Butcher SA, Gurr SJ, Lebrun MH, Ridout CJ, Schulze-Lefert P, Talbot NJ, Ahmadinejad N, Ametz C, Barton GR, Benjdia M, Bidzinski P, Bindschedler LV, Both M, Brewer MT, Cadle-Davidson L, Cadle-Davidson MM, Collemare J, Cramer R, Frenkel O, Godfrey D, Harriman J, Hoede C, King BC, Klages S, Kleemann J, Knoll D, Koti PS, Kreplak J, López-Ruiz FJ, Lu X, Maekawa T, Mahanil S, Micali C, Milgroom MG, Montana G, Noir S, O’Connell RJ, Oberhaensli S, Parlange F, Pedersen C, Quesneville H, Reinhardt R, Rott M, Sacristán S, Schmidt SM, Schön M, Skamnioti P, Sommer H, Stephens A, Takahara H, Thordal-Christensen H, Vigouroux M, Wessling R, Wicker T, Panstruga R (2010) Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330:1543–1546. doi:10.1126/science.1194573
Tamayo E, Gómez-Gallego T, Azcón-Aguilar C, Ferrol N (2014) Genome-wide analysis of copper, iron and zinc transporters in the arbuscular mycorrhizal fungus Rhizophagus irregularis. Front Plant Sci 14(5):547. doi:10.3389/fpls.2014.00547
Tisserant E, Kohler A, Dozolme-Seddas P, Balestrini R, Benabdellah K, Colard A, Croll D, Da Silva C, Gomez SK, Koul R, Ferrol N, Fiorilli V, Formey D, Franken P, Helber N, Hijri M, Lanfranco L, Lindquist E, Liu Y, Malbreil M, Morin E, Poulain J, Shapiro H, van Tuinen D, Waschke A, Azcón-Aguilar C, Bécard G, Bonfante P, Harrison MJ, Küster H, Lammers P, Paszkowski U, Requena N, Rensing SA, Roux C, Sanders IR, Shachar-Hill Y, Tuskan G, Young JP, Gianinazzi-Pearson V, Martin F (2012) The transcriptome of the arbuscular mycorrhizal fungus Glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont. New Phytol 193:755–769. doi:10.1111/j.1469-8137.2011.03948.x
Tisserant E, Malbreil M, Kuo A, Kohler A, Symeonidi A, Balestrini R, Charron P, Duensing N, Frei dit Frey N, Gianinazzi-Pearson V, Gilbert LB, Handa Y, Herr JR, Hijri M, Koul R, Kawaguchi M, Krajinski F, Lammers PJ, Masclaux FG, Murat C, Morin E, Ndikumana S, Pagni M, Petitpierre D, Requena N, Rosikiewicz P, Riley R, Saito K, San Clemente H, Shapiro H, van Tuinen D, Bécard G, Bonfante P, Paszkowski U, Shachar-Hill YY, Tuskan GA, Young JP, Sanders IR, Henrissat B, Rensing SA, Grigoriev IV, Corradi N, Roux C, Martin F (2013) Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc Natl Acad Sci U S A 110:20117–20122. doi:10.1073/pnas.1313452110
Tollot M, Wong Sak Hoi J, van Tuinen D, Arnould C, Chatagnier O, Dumas B, Gianinazzi-Pearson V, Seddas PM (2009) An STE12 gene identified in the mycorrhizal fungus Glomus intraradices restores infectivity of a hemibiotrophic plant pathogen. New Phytol 181:693–707. doi:10.1111/j.1469-8137.2008.02696.x
Trouvelot A, Kough J, Gianinazzi-Pearson V (1986) Evaluation of VA infection levels in root systems. Research for estimation methods having a functional significance, In: Gianinazzi-Pearson V, Gianinazzi S (eds) Physiological and Genetical Aspects of Mycorrhizae. INRA Press, France, p 217–221
Villarreal-Ruiz L, Anderson IC, Alexander IJ (2004) Interaction between an isolate from the Hymenoscyphus ericae aggregate and roots of Pinus and Vaccinium. New Phytol 164:183–192. doi:10.1111/j.1469-8137.2004.01167.x
Young JPW (2015) Genome diversity in arbuscular mycorrhizal fungi. Curr Opin Plant Biol 23:113–119. doi:10.1016/j.pbi.2015.06.005
Acknowledgments
Research was funded by the BIOBIT-Converging Technology project (WP2), the Progetto Ateneo SLEPS and the University grant (60 %) to LL. We thank Maria J. Harrison for the mtpt4-2 mutant, Nuria Ferrol for the RFP yeast strains, Andrea Genre for confocal microscopy observations, Raffaella Balestrini for the help on laser microdissection, and Paola Bonfante and Silvia Perotto for fruitful discussions and critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
(DOCX 12 kb)
Figure S1
Nucleotide and deduced amino acid sequences of RiPEIP1. (a) RiPEIP1 genomic DNA sequence showing the presence of five introns. (b) RiPEIP1 protein sequence showing the 4 transmembrane domains (underlined), the typical ER-retention/retrieval motifs (bold) and the predicted phosphorylation sites (red). See text for details (GIF 255 kb)
Figure S2
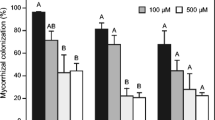
Colonization level of M. truncatula roots at 28 and 60 days post inoculation (dpi) assessed accordingly to Trouvelot et al. (1986). F%: frequency of mycorrhization in the root system (a), a%: arbuscules abundance in mycorrhizal parts of root fragments (b). Different letters indicate statistically significant difference (p < 0.05 ANOVA, Tukey’s post-hoc test) (GIF 17 kb)
Figure S3
Mycorrhizal phenotype of M. truncatula wt (a, b) and mtpt4-2 (c, d) roots colonized by R. irregularis. Roots were harvested 60 dpi, stained with acid fuchsin and observed with a confocal microscope. Arbuscules in mtpt4-2 are degenerated as described in Javot et al. (Javot et al. 2011). Bars = 25 μm. (GIF 119 kb)
Figure S4
Molecular analyses of O. maius transgenic strains expressing RiPEIP1. (a) Gel electrophoresis of PCR products obtained from genomic DNA of wt and transgenic strains using RiPEIP1 specific primers (b) Southern blot of genomic DNA from wt and transgenic strains restricted with BglII enzyme and hybridized with the RiPEIP1 probe. Lanes corresponding to BC6, BA2, BA4 samples exhibited a single genomic insertion. (GIF 114 kb)
Figure S5
Phenotype of O. maius free living mycelia from wt and RiPEIP1-expressing strain BA2 as revealed by calcoflour white staining. Laser-scanning microscope observation of one-month-old O. maius wt and transgenic strains stained with calcofluor white. Transmitted light images are shown on the right (b, d) and the corresponding fluorescence images on the left (a, c). Bars: 10 μm (GIF 188 kb)
Figure S6
Mycorrhizal phenotype of O. maius wt and RiPEIP1-expressing strain BC6. Mycorrhized V. myrtillus roots were stained with acid fuchsin and observed 2 months after fungal inoculation. O. maius transgenic strains showed an increased number of coils (asterisk) in epidermal cells compared to the wt. (GIF 426 kb)
Rights and permissions
About this article
Cite this article
Fiorilli, V., Belmondo, S., Khouja, H.R. et al. RiPEIP1, a gene from the arbuscular mycorrhizal fungus Rhizophagus irregularis, is preferentially expressed in planta and may be involved in root colonization. Mycorrhiza 26, 609–621 (2016). https://doi.org/10.1007/s00572-016-0697-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-016-0697-0