Abstract
The gastric pathogen Helicobacter pylori is one of the most genetically diverse bacteria. Recombination and DNA transfer contribute to its genetic variability and enhance host adaptation. Among the strategies described to increase genetic diversity in bacteria, DNA transfer by conjugation is one of the best characterized. Using this mechanism, a fragment of DNA from a donor cell can be transferred to a recipient, always mediated by a conjugative nucleoprotein complex, which is evolutionarily related to type IV secretion systems (T4SSs). Interestingly, the H. pylori chromosomes can encode up to four T4SSs, including the cagPAI, comB, tfs3, and tfs4 genes, some of which are known to promote chronic H. pylori infection. The T4SS encoded by the cagPAI mediates the injection of the effector protein CagA and proinflammatory signaling, and the comB system is involved in DNA uptake from the environment. However, the role of tfs3 and tfs4 is not yet clear. The presence of a functional XerD tyrosine recombinase and 5′AAAGAATG-3′ border sequences as well as two putative conjugative relaxases (Rlx1 and Rlx2), a coupling protein (TraG), and a chromosomal region carrying a putative origin of transfer (oriT) suggest the existence of a DNA transfer apparatus in tfs4. Moreover, a conjugation-like DNA transfer mechanism in H. pylori has already been described in vitro, but whether this occurs in vivo is still unknown. Some extrachromosomal plasmids and phages are also present in various H. pylori strains. Genetic exchange among plasmids and chromosomes, and involved DNA mobilization events, could explain part of H. pylori’s genetic diversity. Here, we review our knowledge about the possible DNA transfer mechanisms in H. pylori and its implications in bacterial adaptation to the host environment.
Similar content being viewed by others
Introduction
The Gram-negative bacterium Helicobacter pylori is a highly successful pathogen colonizing the surface of the gastric eptithelium in the human stomach. H. pylori is commonly transmitted in early childhood and can persist throughout the entire lifetime of the host due to its ability to adapt to changes in its environmental niche [1, 2]. It is well known that about 50 % of the world population carries H. pylori, which causes asymptomatic or chronic gastritis in all infected individuals, while less often (1–15 %) more severe gastric diseases can appear. The clinical outcome of infection with H. pylori depends on a highly complex scenario of host–pathogen interactions. Several parameters such as bacterial genotype, environmental aspects, and genetic predisposition of the host control disease development and progression. H. pylori infections are responsible for a high rate of morbidity and mortality in the world population as a consequence of peptic ulcer disease, mucosa-associated lymphoid tissue lymphoma (MALT), and the most dangerous complication, gastric adenocarcinoma. Gastric adenocarcinoma represents the second leading cause of cancer-associated death globally, with about 700,000 people dying every year [1, 2]. Due to these cancerogenic activities, H. pylori was the first bacterium to be classified as a type-I carcinogen by the World Health Organization’s International Agency for Research on Cancer.
Phylogenetic and other similarities indicated that H. pylori is approximately as old as are anatomically modern humans, and was acquired about 100,000 years ago in Africa via a single host jump from an unknown non-human host [3]. Today, H. pylori is considered one of the most genetically diverse bacterial pathogens worldwide [4–7]. This diversity can be attributed to altered DNA repair, elevated mutation rates, high frequency of intraspecific recombination, chromosomal DNA transfer events from other species, incorporation of naked DNA from the environment, plasmid mobilization, and integration of acquired DNA in the so-called plasticity zones [4, 8–10]. In addition, the presence of phages has been described for several H. pylori strains, acting as another source of bacterial diversity and evolution [11–15]. Using newly developed bioinformatics programs, recent comparative studies of 29 complete genome sequences of strains from different parts of the world revealed a high-resolution picture of the H. pylori population structure as well as the extent and direction of genetic flux between subgroups of the bacterium [16]. These analyses suggest the occurrence of frequent recombination events and other important changes at the genetic level, which help to explain the organism’s evolution in the stomach and certain adaptation events during transmission [16]. Here we review the different known and proposed mechanisms used by H. pylori to acquire and transfer exogenous DNA, and thus, increase its genetic variability and adaptation to the host (Figure 1). We also provide an overview of the various DNA transfer machineries employed by H. pylori to deliver genetic material to other bacterial species and possibly even to organisms in different kingdoms.
Schematic model for the possible role of type IV secretion systems in DNA transfer events of the gastric pathogen Helicobacter pylori. Each potential DNA transfer mechanism is indicated with the corresponding name and red arrows, which includes the proposed direction of DNA delivery. H. pylori is coloured in green, C. jejuni in brown, and the mammalian host cell in grey
General role of type IV secretion systems (T4SSs) in DNA exchange
T4SSs represent a family of conserved molecular transporters in bacteria that are designed to deliver a variety of molecules [17]. The Agrobacterium tumefaciens VirB/D4 system is the prototypical T4SS, which is formed by 11 VirB proteins (named as VirB1–VirB11) and a coupling protein (called VirD4). These factors have been categorized into three subgroups: (1) cytoplasmatic or inner membrane proteins (three ATPases: VirB4, VirB11 and VirD4), (2) the “core channel component” subunits that constitute the central channel structure (VirB6–VirB10) putting in contact the inner and outer bacterial membranes, and (3) a pilus surface appendage composed of VirB2 and VirB5 proteins, considered as the major and minor pilus subunits, respectively [18]. Although T4SSs are variable with regard to the number of encoded proteins that establish the structure of the secretion channel, most of them are composed of eleven VirB proteins [18]. This fascinating transporter family possesses a high versatility based on the nature of the translocated substrates, which can be DNA, proteins, or nucleoprotein complexes, but also regarding the final destination of the secreted substrates. DNA can be mobilized between bacteria in the conjugation process [19–21] or delivered to eukaryotic cells [22]. However, T4SSs can also be used for the uptake of naked DNA from the environment or DNA release into the extracellular milieu. Therefore, T4SS family members have been classified into three different subfamilies according to their main function [23]: (1) conjugation of chromosomal and plasmid DNA [19–21], (2) effector protein translocation into target cells [24], and (3) DNA uptake and release to the extracellular milieu [25].
Single H. pylori strains can encode up to four different T4SSs in the chromosome: the cytotoxin-associated genes pathogenicity island (cagPAI), comB, tfs3, and tfs4 (Fig. 2). The cagPAI and comB gene systems have been characterized in great detail. The cagPAI is a locus of about 40 kb and contains approximately 30 genes [26]. All orthologs of the 11 VirB proteins have been identified as well as some accessory factors leading to a T4SS model similar to that of A. tumefaciens [27]. The only reported effector protein translocated into the host cell by the cagPAI T4SS is CagA [28]. Therefore, CagA has been recognized as a genetic marker for the cagPAI locus [29] and H. pylori-associated disease development [1, 2]. Sequence comparison of the cagPAI genes of more than 30 H. pylori isolates have shown that the gene order and content are highly conserved [30]. The phylogeny of most cagPAI genes was similar to that of housekeeping genes, indicating that it was acquired by a single event more than 60,000 years ago, and its genetic diversity reflects the isolation by distance that has shaped this bacterial species since modern humans migrated out of Africa [30]. The fact that the 40 kb cagPAI has been incorporated by an ancient DNA transfer episode represents a remarkable example for its importance in acquiring genetic elements for bacterial virulence. In addition, H. pylori uses the ComB T4SS for uptake of naked DNA during natural transformation, a very effective source to gain genetic variability [31], shown to be important for promoting chronic H. pylori infection [32]. The T4SS member tfs3 has also been postulated to have a function in horizontal DNA transfer [33]. Later on, a role in DNA transfer has also been proposed for another chromosomal locus named tfs4 [34]. Although the mechanism of horizontal DNA transfer in H. pylori is widely still unclear, its genetic diversity is a highly remarkable feature [6–10].
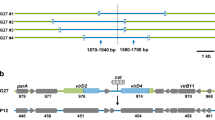
Helicobacter pylori chromosomes can encode for up to four type IV secretion systems. a Schematic presentation of the chromosome of H. pylori strain P12. The two outermost concentric circles denote the predicted coding genes represented as bars on the plus (external blue circle) and minus strand (internal blue circle). The third concentric circle (black peaks) represents the guanine and cytosine (G+C) content. Purple and green peaks in the last concentric circle denote the G+C skew calculated with a window size of 10,000 and steps of 100 bp. This figure was created using the CG View program [92]. The location of four T4SSs (comB, cagPAI, tfs3, and tfs4 genes) is indicated by red bars. The three plasticity zones are represented with orange bars. b Genetic composition of four different T4SSs according to their position in the genome of H. pylori strain P12 (cag genes in the cagPAI, where virB homologues are indicated with red letters, comB genes in the comB T4SS cluster and virB homologue genes in the tfs3 and tfs4 cluster, respectively). Red arrows represent genes which are described to encode structural T4SS components. Blue indicates putative conjugative relaxases; green indicates the coupling proteins, and yellow, the ATPases. The grey genes have yet unknown functions
The ComB system of DNA uptake
A wide variety of Gram-positive and Gram-negative bacteria are competent for natural transformation, being able to import exogenous DNA from the environment and insert it into their chromosomes [35]. While the advantage of this mechanism is still not fully understood, it likely allows pathogenic bacteria to better adapt to their hosts over years and decades of infection. Natural transformation in bacteria is usually correlated with the presence of a type IV pilus-based DNA uptake system as described for Neisseria gonorrhoeae [36], Legionella pneumophila [37], and many others. However, in H. pylori this mechanism is related to a specific T4SS, the ComB system. The ComB apparatus described for H. pylori [38] and Campylobacter jejuni [39] are the only known exceptions that use a T4SS variant to facilitate natural transformation. This is certainly a remarkable alternative to the well studied mechanism used by the type IV pilus-based system. Furthermore, ComB-dependent natural transformation in H. pylori varies in efficiency between strains, but is saturable and sensitive to the length, symmetry, and strandedness of the DNA substrate [40].
The H. pylori ComB apparatus comprises a nearly complete set of T4SS components with a similar gene cluster organization, which were named according to their orthologous proteins of the A. tumefaciens VirB/D4 T4SS. The ComB system only lacks VirB1 and VirB5 factors, as well as the VirB11 ATPase (Table 1; Fig. 2). These three components are also missing in a few T4SSs from other bacteria, and appear to be dispensable in these systems. However, the possibility to find functionally equivalent genes encoding ComB orthologs for VirB1, VirB5, and VirB11 somewhere else in the H. pylori genome cannot be excluded. The comB gene cluster was originally discovered by screening an H. pylori mutant library generated by mini-TnblaM transposon shuttle mutagenesis. One of the mutants was unable to be transformed with plasmid DNA, and transformation with chromosomal DNA was also strongly reduced [38]. The comB locus was first described to comprise the genes comB1 to comB3, and a short open reading frame (ORF) designated as orf2 [38]. By using the BLASTP algorithm, protein homologies of ComB2 and ComB3 to VirB9 and VirB10 of A. tumefaciens were described [38]. Later on, ComB1 was identified as a member of the VirB8 protein family, and ORF2 was predicted to encode a VirB7-like protein containing a putative lipoprotein signal sequence [31]. For a better correlation with the A. tumefaciens VirB components, it was proposed that ComB1, ComB2, and ComB3 should be re-named as ComB8, ComB9, and ComB10, respectively. Interestingly, the periplasmic C-terminal domain of H. pylori ComB10 has been crystallized as a dimer and used as a model for the periplasmic domains of VirB10-like family proteins [41]. In addition, ORF2 is now referred to as ComB7 [31]. Another ORF with significant sequence homology to A. tumefaciens VirB4 ATPase was later designated as ComB4. The corresponding gene is closely located in the vicinity of the comB locus and contains the typical ATP/GTP binding motif of ATPases. A comB4 deletion mutant in H. pylori is deficient for DNA import; however, it is not yet clear whether the ATPase activity is essential for channel assembly and/or to energize the DNA transport [31]. Deletion mutants of comB4, comB8, comB9 and comB10 genes were generated and shown to be absolutely essential for transformation competence in H. pylori, whereas comB7 is not [31]. Later on, by database comparison analysis of individual ORFs surrounding the described comB cluster, three additional genes were identified: comB2, comB3 and comB6 (Table 1) [25]. Deletion of comB2 and comB3 genes abrogated the uptake of natural plasmids and chromosomal transformation. By bioinformatic analysis, it was determined that ComB6 is a polytopic membrane protein containing five to seven transmembrane domains, similar to agrobacterial VirB6. A comB6 deletion mutant was completely defective in plasmid uptake and natural transformation of chromosomal DNA [25]. A topology study, protease digestion experiments, and secondary structure predictions conducted by Hofreuter and co-workers allowed them to define ComB7 as a membrane-attached protein and ComB8 as a membrane-associated protein with a short N-terminal domain in the periplasm [42]. Furthermore, ComB9 was identified as a periplasmic protein attached to the membrane, and the N-terminus of ComB10 seems to be anchored in the cytoplasmic membrane, while the main part of the protein is located in the periplasm [42]. Apart from the ComB system, additional proteins involved at different steps in the transformation competence process have also been identified, such as the cytoplasmatic proteins RecA [43], DprA [44], the secreted protein ComH [45], and the cytoplasmatic channel, ComEC, previously described in Bacillus subtilis [46]. These factors also appear to be essential for DNA uptake in H. pylori.
Based on the homology of H. pylori ComB components to A. tumefaciens VirB proteins, a current model has been postulated where ComB2, ComB3, ComB4, ComB6, ComB8, ComB9, and ComB10 are crucial components for the uptake of DNA [42]. The ComB7 protein seems to stabilize the central channel structure “core complex”, but is dispensable for T4SS functionality [42]. Using a method to localize DNA labeled with a fluorescent marker, a two-step uptake mechanism for transport of external DNA by H. pylori has been proposed [47]. The ComB T4SS was implicated in this process during the outer membrane double-stranded (ds) DNA transport, and the ComEC system seems necessary for uptake of the DNA from the periplasm to the cytoplasm [47]. These data indicate that there are differences in outer and inner membrane DNA import, which await further characterisation. Taken together, it appears that the ComB system has evolved conservatively in all known H. pylori strains. This conservation is in agreement with the need for genomic fluidity, enabling the bacteria to keep the gene content flexible and as diverse as possible to survive in the stomach during the lifelong colonization and infection process. In agreement with this assumption, the comB4 and comB3 genes have been identified in transposon mutant library screens to be essential for colonization in gerbils and mice, respectively [48, 49]. This suggests that natural competence may indeed play a role in host colonization. However, in further studies it was also shown that a comB8–10 mutant strain, which was deficient in natural transformation competence as described above, was still able to colonize gerbils very efficiently in single-infection experiments [48]. From these data it could be suggested that some comB genes, besides a role in natural transformation competence, might have an additional essential function for H. pylori which has not been identified yet.
Conjugation as a general mechanism of DNA transfer
Conjugation is a mechanism of horizontal DNA transfer initially reported to occur between two bacterial cells. Here DNA is transferred from the donor cell to a given recipient, and this transfer is mediated by a protein complex known as the conjugative apparatus. Among all the pathways described to increase bacterial genetic diversity, conjugation is very important in nature, as it constitutes a mechanism that contributes to the transmission of virulence factors and antibiotic resistances [50], which can enhance host adaptation [51]. Under laboratory conditions conjugation has also been observed between organisms from different kingdoms: from bacteria to plants [52], between bacteria and yeast [53], and even from bacteria to mammalian cells [54]. Under natural conditions, DNA transfer between bacteria and eukaryotic plant cells has been well documented for certain species of Agrobacterium. These pathogens can mobilize a DNA segment, known as T-DNA, in a process evolutionarily related to bacterial conjugation [55]. This mechanism has also been recently described for a plasmid construct mobilized from the human pathogen Bartonella henselae to human vascular endothelial cells [56, 57]. In general, DNA transfer by conjugation can be defined as the interplay between two separate processes: (1) a DNA rolling-circle replication (RCR) mechanism mediated by a nucleoprotein complex (relaxase-DNA) and (2) delivery of macromolecules into the recipient cell through a secretion channel, the T4SS [58]. This assumption is based on the fact that conjugative DNA processing enzymes exhibit high sequence similarity with those implicated in RCR. Moreover, their target sequence, the origin of transfer (oriT) and the origin of replication in the RCR, are closely related to each other and share structural similarities [59].
Plasmids in H. pylori
Genetic variability in H. pylori may not only arise by the uptake of free DNA through the ComB system. H. pylori also possesses a conjugation-like mechanism for the DNA exchange between clinical strains [60]. Furthermore, the exchange of plasmids may also contribute to this genetic diversity. The existence of various plasmids in H. pylori was demonstrated for the first time more than 22 years ago [61]. Despite several complete H. pylori genome sequences that are available in public databases, the structure and function of plasmids in these bacteria have not been comprehensively characterized yet. About 50 % of clinical H. pylori strains carry cryptic plasmids ranging in size from 2 to 100 kb [62, 63]; however, their genetic content, gene organization, and function are still not fully understood. The majority of these plasmids have a size of 2 to 6 kb (Table 2). They code for an origin of replication (ori), replication genes (repA or repB), and occasionally, a small ORF with unknown function [62, 63]. In contrast, the larger H. pylori plasmids carry a number of additional ORFs with unknown function (Table 2). All of these plasmids may be classified into two groups. Those that revealed homology to plasmids of Gram-positive bacteria replicating via the RCR mechanism are in group 1, and members of the second group replicate via the “theta” mechanism and carry direct repeat sequences called “iterons” [62].
According to capacity for being mobilizable, plasmids can also be classified as conjugative (transmissible by its own functions), mobilizable (transmissible in the presence of a helper conjugative plasmid), or non-mobilizable. Mobilizable plasmids are usually smaller than conjugative ones, and encode for an oriT and proteins involved in conjugative DNA processing, such as relaxase enzymes, coupling proteins and nicking accessory factors [64]. The remaining part of the conjugative machinery can be provided in trans by a helper plasmid or the bacterial chromosome. On the other hand, conjugative plasmids encode for the whole machinery necessary for their mobilization, the complete T4SS channel, together with the above mentioned proteins involved in the DNA processing and transfer [64]. Several plasmids identified in clinical H. pylori strains seem to encode for typical conjugative mobilization (mob) genes (Table 2). The origin of these H. pylori plasmids and whether they can also replicate in other bacterial species are yet unknown. These important questions should be investigated in future studies.
Evidence for multiple pathways of plasmid transfer in H. pylori
Very effective restriction barriers have been identified in bacteria, which act as a hitch for plasmid transfer by natural transformation. For example, H. pylori encodes a variety of restriction–modification (R-M) systems as a defense strategy against invasion by foreign DNA such as that of phages [65]. To circumvent R-M systems, an alternative method for DNA exchange might be transfer by conjugation. During conjugation, single-stranded (ss) DNA is delivered, which cannot be cleaved by restriction enzymes. Moreover, DNA transfer by conjugation in laboratory experiments should also be resistant to externally given DNaseI, because the DNA is transferred through the T4SS secretion channel from the donor to the recipient cell and is therefore protected.
Among the multitude of plasmids described in various H. pylori clinical strains (Table 2), only some are noted to carry a so-called mob region comprising putative mobilization genes such as relaxases, coupling factors, and possible accessory proteins together with an oriT sequence [34, 66–71]. Characterization of the 10.9 kb plasmid pHel4 led to the identification of the first mobilizable extrachromosomal element encoded in H. pylori [66]. Afterwards, pHel12, another cryptic mobilizable plasmid of 10.2 kb, was also reported [34]. Putative mob regions were identified in the pHel4 and pHel12 plasmids, with similarities to mobA, mobB, mobC, and mobD genes previously described in the Escherichia coli pColA, pColE1, or pColD plasmids. Both H. pylori plasmids also carry a microcin gene cluster, mccC7, like the one present in E. coli plasmid pMccC7 [72]. Interestingly, MccC7 is a linear heptapeptide that is active on bacteria phylogenetically related to E. coli and inhibits protein translation in vivo [73] and in vitro [74]. The fact that H. pylori can carry plasmids with putative mob regions suggests that there is a conjugative mechanism by which these plasmids can be mobilized. Recently, multiple pathways for plasmid DNA transfer in H. pylori have been investigated [63]. The authors proposed that plasmid transfer can proceed by two mechanisms, DNaseI-sensitive natural transformation and a DNaseI-resistant alternative (ADR) transfer. Plasmid transfer frequencies were determined, and deletion of the mobA gene had only a small effect on plasmid mobilization, suggesting that relaxase MobA (encoded in pHel4 and pHel12) ADR is not involved in this function. Thus, it could be that MobA may act in RCR of the plasmid as a replication initiator protein. A possible complementation effect with one of the two chromosomally encoded H. pylori relaxases [75], however, has been excluded by the authors [63]. Interestingly, the two mobilizable plasmids pHel4 and pHel12 do not encode T4SS components (Table 2), implying that one of the various chromosomal T4SSs could be used for plasmid transfer. To study the role of the chromosomal T4SSs in the proposed plasmid DNA transfer, all the T4SSs were sequentially inactivated by mutagenesis. Surprisingly, deletion of single chromosomal T4SSs or even all four T4SSs in the donor strain did not downregulate plasmid transfer rates. However, when the comB gene system is deleted in the recipient strain, plasmid transfer values were reduced in experiments with or without externally given DNaseI, indicating that ComB T4SS is implicated in the uptake of pHel4 and pHel12 plasmids in the recipient by transformation as well as DNaseI-resistant mobilization [63]. Taken together, Rohrer and co-workers provided evidence that H. pylori plasmids pHel4 and pHel12 can be transferred both by conjugation and natural transformation mechanisms [63]. In both cases, the MobA relaxase in the donor and the ComB system in the recipient were strictly necessary. It is remarkable that they have also detected the possibility of DNA mobilization in the absence of any T4SS in the donor, suggesting an alternative way for plasmid mobilization, which is independent of any T4SS and ADR mechanism [63]. In addition to pHel4 and pHel12, conjugative plasmid DNA transfer was shown for pHP69 [69], while it remains unknown for all other mob gene-encoding plasmids (Table 2). However, a couple of important questions remain unanswered. For example, it is still unknown which genes encode the proteins forming the functional conjugative membrane channel. Is this a typical T4SS or different transporter? Since these transfer genes are not encoded by the above discussed plasmids, they should be located in the chromosome. Thus, more work is certainly necessary to characterize this transfer in more detail.
Putative DNA transfer genes encoded in the H. pylori chromosome
Apart from the cagPAI and comB T4SS genes, Kersulyte et al. [33] identified a novel 16.3 kb gene cluster located in one of the three plasticity regions in H. pylori, which encodes a putative third T4SS, named as Tfs3 (Fig. 2). Due to the low G+C content in the tfs3 gene cluster as compared to the rest of the genome, it has been suggested that it was acquired by horizontal DNA transfer from another bacterial species [33]. Tfs3 is composed of 16 ORFs in the first described H. pylori strain, PeCan18B, and seven of them exhibit homology to those previously described for the virB/D4 operon in A. tumefaciens [33]. The functional role of tfs3, however, is still not clear, but some researchers suggested a possible implication in DNA transfer [33]. Interestingly, full-length and partially truncated tfs3 genes were found in 20 and 19 %, respectively, of 94 clinical H. pylori strains tested [33]. Due to the sequence variation and presence of different genes, some authors classified tfs3 into two subgroups, tfs3a and tfs3b [76]. However, it seems that tfs3a and tfs3b are not related to each other. Tfs3b is clearly distinct and has been called Tfs4 (for fourth T4SS) by other groups [34], which may cause some confusion in the literature. In order to have an unequivocal nomenclature and to avoid misunderstandings, we propose to use exclusively the Tfs3 and Tfs4 terminology in the future.
Tfs4 was identified and characterized initially when the complete genome sequence of the H. pylori strain P12 had been published [34]. As described for tfs3, tfs4 genes are located in one of the three plasticity zones of the H. pylori chromosome (Fig. 2) and harbor a complete set of orthologs to constitute a putative T4SS channel [34]. It also includes the disease marker dupA (duodenal ulcer promoting gene A), a virB4 homologue [77]. Interestingly, dupA of strain J99 (also designated jhp0917/jhp0918) was found to be essential for the colonization of H. pylori strains NSH79 and NSH57 in mice, suggesting an important role for the establishment and maintenance of H. pylori infection [49]. Recent work also demonstrated that large fragments of the tfs4 gene cluster can be horizontally transferred among H. pylori isolates in a manner dependent on the XerD protein (Table 3), a tyrosine recombinase encoded within tfs4. XerD excises the tfs4 gene fragment at a specific sequence (5′-AAAGAATG-3′), generating a circular intermediate that is transfered to a given recipient cell via the Tfs4 T4SS [34].
The first notion of a potential conjugative DNA transfer region in H. pylori was reported in the description of a putative TraG/VirD4 coupling protein (HP1006) encoded in Tfs4 of strain 26695, besides the already known VirD4 protein in the cagPAI T4SS (Table 3) [75]. Moreover, two closely associated ORFs (HP0996 and HP1004) with significant homology to conjugative relaxases were also detected and named as Rlx1 and Rlx2, respectively (Table 3) [75]. The homologous relaxase protein in the Agrobacterium VirB/D4 T4SS has been called VirD2 according to its function in T-DNA transfer to plants [17, 18]. However, since it is unknown if either of the two H. pylori relaxases could act similarly to agrobacterial VirD2 and to distinguish between the two enzymes, we propose to name these proteins in all H. pylori strains always with the Rlx1/2 nomenclature. Rlx1 and Rlx2 contain at their N-terminal domain (1–300 aa) the three specific relaxase motifs that typically characterize conjugative relaxase enzymes [78]. Motifs I and II are involved in catalyzing the nicking–closing reaction at the specific DNA transfer sequence, named as oriT. Motif I carries the tyrosine residue that covalently attaches the relaxase to the 5′ terminus of the cleavage ssDNA. Motif II works as a recognition domain, and motif III contains a sequence pattern essential for relaxase activity. The implication of Rlx1, Rlx2, and the TraG-like protein in plasmids construct mobilization between two H. pylori strains was examined in detail using mutagenesis and primer extension studies [79]. Two shuttle plasmids, called pSB13 and pSB14, carrying the oriT from the RP4 (IncPα) plasmid sequence can be mobilized in a conjugative manner between two H. pylori strains. This conjugative transfer mechanism was dependent on the Rlx1 and the TraG-like protein, but independent of the comB, the cagPAI, and tfs3 genes in the donor [79]. Rlx2 [75], also called VirD2-like protein in another study [80], is encoded in the tfs4 gene cluster. In vitro characterization has shown that Rlx2 can specifically recognize the nicking sequence 5′-ATCCTG-3′ [80]. It is also considered that Rlx2 might function to initiate transfer or XerD-excised tfs4 intermediates [80]. Moreover, a putative oriT region encoded upstream of the rlx2 gene and a VirC1-like relaxosome protein have also been identified (Table 3) [80]. All these data suggest that the tfs4 gene cluster may encode a self transmissible conjugative apparatus; however, there is still no in vivo evidence for such DNA transfer.
Studies of chromosomal DNA transfer among H. pylori strains and from H. pylori to other bacteria
In 1998, Kuipers and co-workers showed for the first time the possibility of a conjugation-like chromosomal DNA exchange mechanism between H. pylori strains in vitro [60]. They have used two H. pylori strains coding either for the resistance against streptomycin (StrepR) or spectinomycin (SpecR), respectively. Conjugation assays were performed on agar plates or in broth medium, in the absence or presence of DNaseI and StrepR, and SpecR double-resistant transconjugands were always obtained [60]. Examination of the progeny by fingerprinting and other genetic methods indicated that the DNA transfer was bidirectional. Mating experiments in the presence of DNaseI reduced but did not eliminate transfer; only the presence of both DNaseI and a membrane separating the cells did so [60]. For progeny derived from matings in the presence of DNaseI, antibiotic resistance and fingerprint profiles indicated that transfer was unidirectional. DNaseI-treated cell-free supernatants also did not transform, ruling out transduction [60]. These experiments indicate that both a DNase-sensitive mechanism (transformation) and a DNase-resistant conjugation-like mechanism involving tight cell-to-cell contact may contribute to chromosomal DNA transfer between two given H. pylori strains. However, it remained unknown if any of the T4SSs is involved in this DNA transfer.
There were also attempts in laboratory experiments to investigate if H. pylori can transfer chromosomal DNA to other bacterial species. Although the natural habitats for H. pylori and Campylobacter jejuni in the gastrointestinal tract are different, the fact that H. pylori can transfer chromosomal DNA (carrying the StrepR marker) to C. jejuni when incubated together on agar plates has been demonstrated [81]. This chromosomal DNA transfer was unidirectional, always from H. pylori into C. jejuni. The transfer was also shown to be independent of the comB, cagPAI and the tfs3 genes in the donor. Moreover, the presence of DNaseI in the medium did not affect the DNA transfer significantly [81]. Although not tested in this report, it may be possible that tfs4 genes could be involved in this DNA transfer.
Conclusions and outlook
Numerous studies have shown that high genetic variability is a remarkable feature of the gastric pathogen H. pylori. Sequence analyses indicated that anatomically modern humans were already infected with H. pylori before their migration out of Africa about 60,000 years ago and demonstrate that this bacterium has remained intimately associated with its human host populations ever since [3]. Genetic diversity occurred within the various H. pylori strains during this long time of co-existence with humans, which helped it to adapt perfectly to its colonization niche. This high genetic variability stimulated the scientific community to investigate how DNA can be gained and lost during bacterial evolution, and several mechanisms have been proposed (Fig. 1). One of the molecular mechanisms that H. pylori used to gain exogenous DNA is by natural transformation using the ComB system. Nevertheless, an increasing number of reports on natural plasmids in H. pylori strains, together with the presence of putative conjugative genes, encouraged researchers to propose that active DNA transfer would play an important role in generating genetic diversity of H. pylori, in addition to other mechanisms. Remarkably, up to four different T4SSs have been described to be encoded in H. pylori chromosomes. Besides the well-described cagPAI and ComB systems, the Tfs3 and Tfs4 T4SSs are located in two different plasticity zones and raise more and more interest. Although their function is still not clear, there is growing evidence for a possible role in DNA transfer. It was shown for the first time that the tfs4 gene island is capable of self-excision and horizontal transfer by a conjugative process, and that DNA excision is mediated by a protein of the XerD family of tyrosine recombinases [34]. In addition, the fact that a putative conjugative relaxase (Rlx2), a coupling protein (TraG-like), and a region proposed to encode the Rlx2-oriT were located just beside the tfs4 genes led us to suggest that these elements may also play a role in a possible conjugative transfer using a relaxase–DNA complex [75]. This assumption should be tested in more detail in future experiments. By comparison, in other human pathogens such as Neisseria gonorrhoeae, ssDNA can be secreted into the extracellular environment, which has a proposed role in biofilm formation of these bacteria [82]. However, if ssDNA export in H. pylori plays a role in colonization in the stomach is yet unknown. Western blotting screens have also shown that Rlx1 and Rlx2 orthologs are possibly expressed in H. acinonychis, a close relative of H. pylori, suggesting that they may be also involved in DNA transfer of other closely related species [83]. However, there are many open questions which await clarification. For example, how do the different proposed DNA transfer systems work in H. pylori? Are relaxases Rlx1 and Rlx2 employed in nicking and delivering of chromosomal DNA, and can these enzymes recognize the oriT sequences identified in the chromosome and mobilizable plasmids? It would also be exciting to find out if H. pylori could transfer DNA to its eukaryotic host, similarly as previously shown for other T4SS-expressing bacteria including A. tumefaciens and B. henselae [56, 57]. It therefore appears that the study of various DNA transfer processes in H. pylori will continue to be a fascinating and rewarding research subject in the future.
References
Atherton JC, Blaser MJ. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J Clin Invest. 2009;119(9):2475–87.
Polk DB, Peek RM. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10(6):403–14.
Moodley Y, et al. Age of the association between Helicobacter pylori and man. PLoS Pathog. 2012;8(5):e1002693.
Alm RA, Trust TJ. Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes. J Mol Med (Berl). 1999;77(12):834–46.
Salama N, et al. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc Natl Acad Sci USA. 2000;97(26):14668–73.
Israel DA, et al. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc Natl Acad Sci USA. 2001;98(25):14625–30.
Suerbaum S, Achtman M. Helicobacter pylori: recombination, population structure and human migrations. Int J Med Microbiol. 2004;294(2–3):133–9.
Kersulyte D, et al. Helicobacter pylori’s plasticity zones are novel transposable elements. PLoS One. 2009;4(9):e6859.
Suerbaum S, et al. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95(21):12619–24.
Gressmann H, et al. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 2005;1(4):e43.
Lehours P, et al. Genome sequencing reveals a phage in Helicobacter pylori. MBio. 2011;2(6):e00239-11.
Heintschel von Heinegg E, et al. Characterisation of a Helicobacter pylori phage (HP1). J Med Microbiol. 1993;38(4):245–9.
Schmid EN, et al. Bacteriophages in Helicobacter (Campylobacter) pylori. J Med Microbiol. 1990;32(2):101–4.
Luo CH, et al. Genome, integration, and transduction of a novel temperate phage of Helicobacter pylori. J Virol. 2012;86(16):8781–92.
Uchiyama J, et al. Complete genome sequences of two Helicobacter pylori bacteriophages isolated from Japanese patients. J Virol. 2012;86(20):11400–1.
Yahara K, et al. Chromosome painting in silico in a bacterial species reveals fine population structure. Mol Biol Evol. 2013;30(6):1454–64.
Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73(4):775–808.
Christie PJ, et al. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta. 2014. doi:10.1016/j.bbamcr.2013.12.019.
Grohmann E, et al. Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67(2):277–301.
Schroder G, Lanka E. The mating pair formation system of conjugative plasmids-A versatile secretion machinery for transfer of proteins and DNA. Plasmid. 2005;54(1):1–25.
Goessweiner-Mohr N, et al. Conjugative type IV secretion systems in gram-positive bacteria. Plasmid. 2013;70(3):289–302.
Llosa M, et al. New perspectives into bacterial DNA transfer to human cells. Trends Microbiol. 2012;20(8):355–9.
Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 2006;9(2):207–17.
Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;1(2):137–49.
Karnholz A, et al. Functional and topological characterization of novel components of the comB DNA transformation competence system in Helicobacter pylori. J Bacteriol. 2006;188(3):882–93.
Covacci A, et al. Helicobacter pylori virulence and genetic geography. Science. 1999;284(5418):1328–33.
Backert S, et al. VirB2 and VirB5 proteins: specialized adhesins in bacterial type-IV secretion systems? Trends Microbiol. 2008;16(9):409–13.
Kwok T, et al. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449(7164):862–6.
Backert S, Selbach M. Role of type IV secretion in Helicobacter pylori pathogenesis. Cell Microbiol. 2008;10(8):1573–81.
Olbermann P, et al. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet. 2010;6(8):e1001069.
Hofreuter D, et al. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol Microbiol. 2001;41(2):379–91.
Dorer MS, et al. Natural competence promotes Helicobacter pylori chronic infection. Infect Immun. 2013;81(1):209–15.
Kersulyte D, et al. Cluster of type IV secretion genes in Helicobacter pylori’s plasticity zone. J Bacteriol. 2003;185(13):3764–72.
Fischer W, et al. Strain-specific genes of Helicobacter pylori: genome evolution driven by a novel type IV secretion system and genomic island transfer. Nucleic Acids Res. 2010;38(18):6089–101.
Solomon JM, Grossman AD. Who’s competent and when: regulation of natural genetic competence in bacteria. Trends Genet. 1996;12(4):150–5.
Biswas GD, et al. Factors affecting genetic transformation of Neisseria gonorrhoeae. J Bacteriol. 1977;129(2):983–92.
Stone BJ, Kwaik YA. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J Bacteriol. 1999;181(5):1395–402.
Hofreuter D, et al. Natural competence for DNA transformation in Helicobacter pylori: identification and genetic characterization of the comB locus. Mol Microbiol. 1998;28(5):1027–38.
Bacon DJ, et al. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect Immun. 2000;68(8):4384–90.
Levine SM, et al. Plastic cells and populations: DNA substrate characteristics in Helicobacter pylori transformation define a flexible but conservative system for genomic variation. FASEB J. 2007;21(13):3458–67.
Terradot L, et al. Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori. Proc Natl Acad Sci USA. 2005;102(12):4596–601.
Hofreuter D, et al. Topology and membrane interaction of Helicobacter pylori ComB proteins involved in natural transformation competence. Int J Med Microbiol. 2003;293(2–3):153–65.
Schmitt W, et al. Cloning of the Helicobacter pylori recA gene and functional characterization of its product. Mol Gen Genet. 1995;248(5):563–72.
Smeets LC, et al. The dprA gene is required for natural transformation of Helicobacter pylori. FEMS Immunol Med Microbiol. 2000;27(2):99–102.
Smeets LC, et al. comH, a novel gene essential for natural transformation of Helicobacter pylori. J Bacteriol. 2000;182(14):3948–54.
Yeh YC, et al. Characterization of a ComE3 homologue essential for DNA transformation in Helicobacter pylori. Infect Immun. 2003;71(9):5427–31.
Stingl K, et al. Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc Natl Acad Sci USA. 2010;107(3):1184–9.
Kavermann H, et al. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J Exp Med. 2003;197(7):813–22.
Baldwin DN, et al. Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect Immun. 2007;75(2):1005–16.
Mazel D, Davies J. Antibiotic resistance in microbes. Cell Mol Life Sci. 1999;56(9–10):742–54.
Ziebuhr W, et al. Evolution of bacterial pathogenesis. Cell Mol Life Sci. 1999;56(9–10):719–28.
Buchanan-Wollaston V, et al. The mob and oriT mobilization functions of bacterial plasmid promote its transfer to plants. Nature. 1987;328:172–5.
Heinemann JA, Sprague GF. Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989;340(6230):205–9.
Waters VL. Conjugation between bacterial and mammalian cells. Nat Genet. 2001;29(4):375–6.
Lessl M, Lanka E. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell. 1994;77(3):321–4.
Fernandez-Gonzalez E, et al. Transfer of R388 derivatives by a pathogenesis-associated type IV secretion system into both bacteria and human cells. J Bacteriol. 2011;193(22):6257–65.
Schroder G, et al. Conjugative DNA transfer into human cells by the VirB/VirD4 type IV secretion system of the bacterial pathogen Bartonella henselae. Proc Natl Acad Sci USA. 2011;108(35):14643–8.
Llosa M, et al. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol. 2002;45(1):1–8.
Waters VL, Guiney DG. Processes at the nick region link conjugation, T-DNA transfer and rolling circle replication. Mol Microbiol. 1993;9(6):1123–30.
Kuipers EJ, et al. Evidence for a conjugation-like mechanism of DNA transfer in Helicobacter pylori. J Bacteriol. 1998;180(11):2901–5.
Kleanthous H, et al. Characterization of a plasmid from Helicobacter pylori encoding a replication protein common to plasmids in gram-positive bacteria. Mol Microbiol. 1991;5(10):2377–89.
Hofler C, et al. Cryptic plasmids in Helicobacter pylori: putative functions in conjugative transfer and microcin production. Int J Med Microbiol. 2004;294(2–3):141–8.
Rohrer S, et al. Multiple pathways of plasmid DNA transfer in Helicobacter pylori. PLoS One. 2012;7(9):e45623.
De la Cruz F, et al. Conjugative DNA metabolism in gram-negative bacteria. FEMS Microbiol Rev. 2010;34(1):18–40.
Ando T, et al. Restriction-modification systems may be associated with Helicobacter pylori virulence. J Gastroenterol Hepatol. 2010;25(Suppl 1):S95–8.
Hofreuter D, Haas R. Characterization of two cryptic Helicobacter pylori plasmids: a putative source for horizontal gene transfer and gene shuffling. J Bacteriol. 2002;184(10):2755–66.
Oh JD, et al. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc Natl Acad Sci USA. 2006;103(26):9999–10004.
Baltrus DA, et al. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol. 2009;191(1):447–8.
Joo JS, et al. Genetic organization and conjugal plasmid DNA transfer of pHP69, a plasmid from a Korean isolate of Helicobacter pylori. J Microbiol. 2012;50(6):955–61.
Carpenter BM, et al. Expanding the Helicobacter pylori genetic toolbox: modification of an endogenous plasmid for use as a transcriptional reporter and complementation vector. Appl Environ Microbiol. 2007;73(23):7506–14.
Xiang Z, et al. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect Immun. 1995;63(1):94–8.
Gonzalez-Pastor JE, et al. Structure and organization of plasmid genes required to produce the translation inhibitor microcin C7. J Bacteriol. 1995;177(24):7131–40.
Garcia-Bustos JF, et al. Structure and mode of action of microcin 7, an antibacterial peptide produced by Escherichia coli. Antimicrob Agents Chemother. 1985;27(5):791–7.
Guijarro JI, et al. Chemical structure and translation inhibition studies of the antibiotic microcin C7. J Biol Chem. 1995;270(40):23520–32.
Backert S, et al. Potential role of two Helicobacter pylori relaxases in DNA transfer? Mol Microbiol. 1998;30(3):673–4.
Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 2010;7(11):629–41.
Lu H, et al. Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology. 2005;128(4):833–48.
Pansegrau W, Lanka E. Enzymology of DNA transfer by conjugative mechanisms. Prog Nucleic Acid Res Mol Biol. 1996;54:197–251.
Backert S, et al. Conjugative plasmid DNA transfer in Helicobacter pylori mediated by chromosomally encoded relaxase and TraG-like proteins. Microbiology. 2005;151(Pt 11):3493–503.
Grove JI, et al. Site-specific relaxase activity of a VirD2-like protein encoded within the tfs4 genomic island of Helicobacter pylori. J Biol Chem. 2013;288(37):26385–96.
Oyarzabal OA, et al. Conjugative transfer of chromosomally encoded antibiotic resistance from Helicobacter pylori to Campylobacter jejuni. J Clin Microbiol. 2007;45(2):402–8.
Zweig MA, et al. Secreted single-stranded DNA is involved in the initial phase of biofilm formation by Neisseria gonorrhoeae. Environ Microbiol. 2013;. doi:10.1111/1462-2920.12291.
Tegtmeyer N, et al. Electron microscopic, genetic and protein expression analyses of Helicobacter acinonychis strains from a Bengal tiger. PLoS One. 2013;8(8):e71220.
Tomb JF, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388(6642):539–47.
Heuermann D, Haas R. Genetic organization of a small cryptic plasmid of Helicobacter pylori. Gene. 1995;165(1):17–24.
Quinones M, et al. Sequence and gene expression analyses of plasmid pHPM8 from Helicobacter pylori reveal the presence of two operons with putative roles in plasmid replication and antibiotic activity. Plasmid. 2001;46(3):223–8.
Minnis JA, et al. Characterization of a 3.5-kbp plasmid from Helicobacter pylori. Plasmid. 1995;34(1):22–36.
De Ungria MC, et al. A novel method of extracting plasmid DNA from Helicobacter species. Helicobacter. 1998;3(4):269–77.
Song JY, et al. Characterization of a small cryptic plasmid, pHP51, from a Korean isolate of strain 51 of Helicobacter pylori. Plasmid. 2003;50(2):145–51.
Hosaka Y, et al. Characterization of pKU701, a 2.5-kb plasmid, in a Japanese Helicobacter pylori isolate. Plasmid. 2002;47(3):193–200.
Song JY, et al. pHP489, a Helicobacter pylori small cryptic plasmid, harbors a novel gene coding for a replication initiation protein. Plasmid. 2003;50(3):236–41.
Grant JR, Stothard P. The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36(Web Server issue):W181–4.
Acknowledgments
We thank PD Dr. Wolfgang Fischer (Max von Pettenkofer Institute, LMU Munich, Germany) for critical comments on the manuscript. The work of SB is supported through a DFG grant (project B10 of CRC-796).
Conflict of interests
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fernandez-Gonzalez, E., Backert, S. DNA transfer in the gastric pathogen Helicobacter pylori . J Gastroenterol 49, 594–604 (2014). https://doi.org/10.1007/s00535-014-0938-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00535-014-0938-y