Abstract
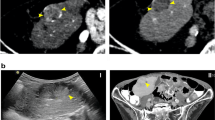
Allelic imbalance (AI), which represents certain chromosomal gains or losses, has been described in hepatocellular carcinoma (HCC), but the significance of AI analysis in focal nodular hyperplasia (FNH) has not been fully clarified. We hypothesized, therefore, that comprehensive allelotyping of FNH could be a useful tool for differentiating FNH from HCC. A 27-year-old man was admitted to the hospital because of general fatigue. A computed tomography (CT) scan disclosed a hepatic nodule 8 cm in diameter. No definite diagnosis was made after imaging or by biopsy before surgery. Macroscopically and microscopically, the surgical specimen showed typical features of FNH. Comprehensive microsatellite analysis was carried out with 382 microsatellite markers distributed throughout all chromosomes. To detect AI effectively, the cutoff value of the AI index was set at 0.70. Among the 382 microsatellite markers, 212 loci were informative, but no AI was detected. The absence of gross chromosomal alterations strongly suggested that the large nodule was FNH rather than HCC, in terms of its genetic background. The patient's subsequent clinical course revealed the nodule to be benign. The results suggest that this genome-wide microsatellite analysis is a useful tool for the differential diagnosis of non-neoplastic liver nodules from HCC.
Similar content being viewed by others
References
BN Nguyen JF Flejou B Terris J Belghiti C Degott (1999) ArticleTitleFocal nodular hyperplasia of the liver. A comprehensive pathologic study of 305 lesions and recognition of new histologic forms Am J Surg Pathol 23 1441–54 Occurrence Handle10584697 Occurrence Handle1:STN:280:DC%2BD3c%2FkvFeqsA%3D%3D Occurrence Handle10.1097/00000478-199912000-00001
T Nishimura N Nishida T Itoh M Kuno M Minata T Komeda et al. (2002) ArticleTitleComprehensive allelotyping of well-differentiated human hepatocellular carcinoma with semiquantitative determination of chromosomal gain or loss Genes Chromosom Cancer 35 329–39 Occurrence Handle12378527 Occurrence Handle1:CAS:528:DC%2BD38XptVGisrk%3D Occurrence Handle10.1002/gcc.10126
T Nishimura N Nishida T Komeda Y Fukuda K Nakao (2005) ArticleTitleGenotype stability and clonal evolution of hepatocellular carcinoma assessed by autopsy-based genome-wide microsatellite analysis Cancer Genet Cytogenet 161 164–9 Occurrence Handle16102588 Occurrence Handle1:CAS:528:DC%2BD2MXos1Wrtrg%3D Occurrence Handle10.1016/j.cancergencyto.2005.02.011
G Wang Y Zhao X Liu L Wang C Wu W Zhang et al. (2001) ArticleTitleAllelic loss and gain, but not genomic instability, as the major somatic mutation in primary hepatocellular carcinoma Genes Chromosom Cancer 31 221–7 Occurrence Handle11391792 Occurrence Handle10.1002/gcc.1138
F Niketeghad HJ Decker WH Caselmann P Lund F Geissler HP Dienes et al. (2001) ArticleTitleFrequent genomic imbalances suggest commonly altered tumour genes in human hepatocarcinogenesis Br J Cancer 85 697–704 Occurrence Handle11531255 Occurrence Handle1:CAS:528:DC%2BD3MXns1yktrw%3D Occurrence Handle10.1054/bjoc.2001.1963
K Shiraishi K Okita N Kusano T Harada S Kondoh S Okita et al. (2001) ArticleTitleA comparison of DNA copy number changes detected by comparative genomic hybridization in malignancies of the liver, biliary tract, and pancreas Oncology 60 151–61 Occurrence Handle11244331 Occurrence Handle1:CAS:528:DC%2BD3MXit1ekt7g%3D Occurrence Handle10.1159/000055313
F Canzian R Salovaara A Hemminki P Kristo PB Chadwick LA Aaltonen et al. (1996) ArticleTitleSemiautomated assessment of loss of heterozygosity and relationship error in tumors Cancer Res 56 3331–7 Occurrence Handle8764130 Occurrence Handle1:CAS:528:DyaK28Xktlaqt7k%3D
IR Wanless C Mawdsley R Adams (1985) ArticleTitleOn the pathogenesis of focal nodular hyperplasia of the liver Hepatology 5 1194–200 Occurrence Handle4065824 Occurrence Handle1:STN:280:BimD2MjktlM%3D
D Cherqui A Rahmouni F Charlotte H Boulahdour JM Metreau M Meignan et al. (1995) ArticleTitleManagement of focal nodular hyperplasia and hepatocellular adenoma in young women: a series of 41 patients with clinical, radiological and pathology correlations Hepatology 22 1674–81 Occurrence Handle7489973 Occurrence Handle1:STN:280:BymD1cfot1U%3D Occurrence Handle10.1016/0270-9139(95)90190-6
YJ Chen PJ Chen MC Lee SH Yeh MT Hsu CH Lin (2002) ArticleTitleChromosomal analysis of hepatic adenoma and focal nodular hyperplasia by comparative genomic hybridization Genes Chromosom Cancer 35 138–43 Occurrence Handle12203777 Occurrence Handle1:CAS:528:DC%2BD38XntVyjt7g%3D Occurrence Handle10.1002/gcc.10103
Author information
Authors and Affiliations
About this article
Cite this article
Nakayama, S., Kanbara, Y., Nishimura, T. et al. Genome-wide microsatellite analysis of focal nodular hyperplasia: a strong tool for the differential diagnosis of non-neoplastic liver nodule from hepatocellular carcinoma. J Hepatobiliary Pancreat Surg 13, 416–420 (2006). https://doi.org/10.1007/s00534-006-1090-8
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00534-006-1090-8