Abstract
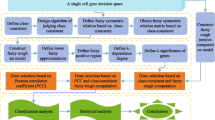
Gene selection is an important technique to remove irrelevant genes and handle the problem of curse-of-dimensionality issue. In other words the objective of the gene selection problem is to find (a small number of) cancer responsible genes (called biomarkers) from large number of genes, which have highest class discernable property. Traditional gene selection techniques are often not scalable on large number of genes and they are not able to handle the problem of vagueness, indiscerniblity, ambiguity, overlappiness complex cancer subtypes classes as usually present in the microarray gene expression data. In this context, a novel greedy fuzzy vaguely quantified rough approach for feature (gene) selection (GFVQRFS) is proposed that handles curse-of-dimensionality issue, vagueness, indiscerniblity, ambiguity, overlapping and complex cancer subtypes classes. The proposed method is evaluated on eight publicly available microarray gene expression datasets and the results are compared with four other state-of-the-art methods namely, CFS-GA, CON-GA, CON-GS and FRFS-GA using three classifiers (viz., KNN, SVM and NB). Six different validity measures (viz., accuracy, precision, recall, macro average \(F_1\)-measures, micro average \(F_1\)-measures and kappa) are used to access the performance of the proposed GFVQRFS method with respect to the compared methods. The proposed method selects very less number of genes compared to those selected by the other counterpart methods. The experimental results reveal the edge of the proposed method over other counterpart methods for most of the datasets.


Similar content being viewed by others
Data availability
Enquiries about data availability should be directed to the authors.
References
Abeel T, Helleputte T, de Peer Y, Dupont P, Saeys Y (2010) Robust biomarker identification for cancer diagnosis with ensemble feature selection methods. Bioinformatics 26(3):392–398
Aha DW, Kibler D, Albert MK (1991) Instance-based learning algorithms. Mach Learn 6:37–66
Alizadeh A, Eisen M, Davis R, Ma C, Lossos I, Rosenwald A, Boldrick J, Sabet H, Tran T, Yu X, Powell J, Yang L, Marti G, Moore T, Hudson J, Lu L, Lewis D, Tibshirani R, Sherlock G, Chan W, Greiner T, Weisenburger D, Armitage J, Warnke R, Levy R, Wilson W, Grever M, Byrd J, Botstein D, Brown P, Staudt L (2000) Distinct types of diffuse large b-cell lymphoma identified by gene expression profiling. Nature 403(6769):503–511
Alon U, Barkai N, Notterman D, Gish K, Ybarra S, Mack D, Levine A (1999) Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Natl. Acad. Sci. 96:6745–6750
Cohen J (1960) A coefficient of agreement for nominal scales. Educ Psychol Measur 20(1):37–46
Dettling M (2004) BagBoosting for tumor classification with gene expression data. Bioinformatics 20(18):583–593
Du D, Li K, Li X, Fei M (2014) A novel forward gene selection algorithm for microarray data. Neurocomputing 133:446–458
Fawcett T (2006) An introduction to ROC analysis. Pattern Recogn Lett 27(8):861–874
Gao K, Khoshgoftaar TM, Napolitano A (2015) An empirical investigation of combining filter-based feature subset selection and data sampling for software defect prediction. Int J Reliab, Qual Saf Eng 22(6):1550027
Golub T, Slonim D, Tamayo P, Huard C, Gaasenbeek M, Mesirov J, Coller H, Loh M, Downing J, Caligiuri M, Bloomfield C (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286(5439):531–537
Halder A, Kumar A (2019) Active learning using rough fuzzy classifier for cancer prediction from microarray gene expression data. J Biomed Inform 92:103136
Halder A, Ghosh S, Ghosh A (2013) Aggregation pheromone metaphor for semi-supervised classification. Pattern Recogn 46(8):2239–2248
Hall MA (1999) Correlation-based feature selection for machine learning. Ph.D. Thesis, The University of Waikato, Hamilton, New Zealand
Jensen R, Cornelis C (2011) Fuzzy-rough nearest neighbour classification and prediction. Theoret Comput Sci 412(42):5871–5884
Jensen R, Shen Q (2009) A new approaches to fuzzy-rough feature selection. IEEE Trans Fuzzy Syst 17(4):310–319
Khan J, Wei J, Ringner M, Saal L, Ladanyi M, Westermann F, Berthold F, Schwab M, Antonescu C, Peterson C, Meltzer P (2001) Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med 6(7):673–679
Kreyszig E (1970) Introductory mathematical statistics, 1st edn. Wily, Hoboken
Kumar A, Halder A (2019) Active learning using fuzzy-rough nearest neighbor classifier for cancer prediction from microarray gene expression data. Int J Pattern Recog Artif Intell 34(1):2057001
Kumar A, Halder A (2020) Ensemble-based active learning using fuzzy-rough approach for cancer sample classification. Eng Appl Artif Intell 91:103591
Liu H, Setiono R (1996) A probabilistic approach to feature selection - a filter solution. In: 13th international conference on machine learning. pp 319–327
Lu Y, Han J (2003) Cancer classification using gene expression data. Inform Syst, Spec issue: Data Manag bioinform 28(4):243–268
Maji P, Pal S (2007) RFCM: a hybrid clustering algorithm using rough and fuzzy sets. Fund Inform 80(4):475–496
Maroulis D, Flaounas I, Iakovidis D, Karkanis S (2006) Microarray-MD: a system for exploratory analysis of microarray gene expression data. Comput Methods Programs Biomed 83(2):157–167
Maulik U, Chakraborty D (2014) Fuzzy preference based feature selection and semisupervised SVM for cancer classification. IEEE Trans NanoBiosci 13(2):1146–1156
Pawlak Z (1991) Rough sets, vol 9 of Theory and Decision Library. Springer, Netherlands
Pawlak Z (1982) Rough sets. Int J Comput Inform Sci 11(5):341–356
Platt JC (1998) Fast training of support vector machines using sequential minimal optimization. In: Schoelkopf B, Burges CJC, Smola AJ (eds) Advances in Kernel methods - support vector learning. The MIT Press, USA, pp 185–208
Singh D, Febbo PG, Ross K, Jackson DG, Manola J, add C, Tamayo P, Renshaw AA, D’Amico AV, Richie JP (2002) Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1(2):203–209
Stekel D (2003) Microarray Bioinformatics, 1st edn. Cambridge University Press, Cambridge, UK
Sun Y, Todorovic S, Goodison S (2010) Local-learning-based feature selection for high-dimensional data analysis. IEEE Trans Pattern Anal Mach Intell 32(9):1610–1626
Tan P, Tan S, Lim C, Khor S (2011) A modified two-stage SVM-RFE model for cancer classification using microarray data. In: Lu B, Zhang L, Kwok J (eds) Neural Information Processing, vol 7062 of Lecture Notes in Computer Science. Springer, Berlin, Heidelberg, pp 668–675
Technology Agency for Science and Research(2022). Kent ridge bio-medical dataset repository. http://datam.i2r.astar.edu.sg/datasets/krbd/index.html
Tou J, Gonzalez R (1977) Pattern recognition principles, 2nd edn. Addison-Wesley, Massachusetts
Tukey JW (1977) Exploratory data analysis. Behavioral Science: Quantitative Methods. Addison-Wesley, Reading, Mass
Wang S, Tang J, Liu H (2016) Feature selection. In: Sammut C, Webb G (eds.), Encyclopedia of machine learning and data mining, Springer US, 2nd edition, pp 1–9
Wei D, Li S, Tan M (2012) Graph embedding based feature selection. Neurocomputing 93:115–125
Zadeh L (1965) Fuzzy sets. Inf Control 8(3):338–353
Funding
Authors declare that this article is not funded by any organization/institute/funding agency.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Conflict of interest declared NONE by the authors.
Research involving human participants and/or animals
Publically available datasets are used for the experiments. No human/ animals are directly involved.
Ethical approval
Authors declare that this article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kumar, A., Halder, A. Greedy fuzzy vaguely quantified rough approach for cancer relevant gene selection from gene expression data. Soft Comput 26, 13567–13581 (2022). https://doi.org/10.1007/s00500-022-07312-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-022-07312-4