Abstract
Automatic segmentation of breast cancer lesions in dynamic contrast-enhanced magnetic resonance imaging is challenged by low accuracy of delineation of the infiltration area, variable structure and shapes, large intensity heterogeneity changes, and low boundary contrast. This study constructed a two-stage breast cancer image segmentation framework and proposes a novel breast cancer lesion segmentation model (TR-IMUnet). The benchmark U-Net network model enables a rough delineation of the breast area in the acquired images and eliminates the influence of unrelated tissues (chest muscle, fat, and heart) on breast tumor segmentation. Based on the extracted results of the region of interest, the rectified linear unit (ReLU) function of the encoding–decoding structure in the model was replaced by an improved ReLU function to reserve and adjust the data dynamically according to input information. The segmentation accuracy of breast cancer lesions was improved by embedding a multi-scale fusion block and a transformer module in the coding path of the model, thereby obtaining multi-scale and global attention information. The experimental results showed that the breast tumor segmentation indexes Dice coefficient (Dice), Intersection over Union (IoU), Sensitivity (SEN), and Positive Predictive Value (PPV) increased by 4.27, 5.21, 3.37, and 3.68%, respectively, relative to the U-Net reference model. The proposed model improves the segmentation results of breast cancer lesions and reduces small area mis-segmentation and calcification segmentation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Breast cancer is a life-threatening disease that can seriously affects both the physical and mental health of patients. Compared with imaging modalities such as molybdenum target, ultrasound, X-ray, magnetic resonance imaging (MRI), and thermography, and other types of imaging modalities (Litjens et al. 2017), DCE-MRI (dynamic contrast-enhanced MRI) is non-invasive and enables visualization of the microvascular network. DCE-MRI is extremely sensitive in detecting early breast cancer, and thus is routinely utilized in clinical practice to assess the local extent of disease and determine the molecular type (Xiao et al. 2021). Automated segmentation of the lesion area in DCE-MRI images has great clinical research value for the early diagnosis, prevention, postoperative guidance, and prognosis of breast cancer; it is the key basis for the development and implementation of artificial intelligence computer-aided diagnosis systems.
In general, DCE-MRI breast cancer types are mainly classified as either mass or non-mass types. Breast cancer lesions are mainly distributed along the gland. The mass type of breast cancer is relatively easy to identify and segment, whereas the non-mass varies in morphology. Cavities, weak boundaries, and low contrast lesion intensity can easily lead to delayed or misdiagnosis, and inadequate treatment. Moreover, manual delineation of the breast cancer lesion area is a very demanding and skillful process, and it is thus necessary for radiologists to have rich clinical experience. In fact, the delineation process can be easily affected by subjectivity, psychological fluctuations, and physical fatigue, leading to random breast lesion delineation results. Consequently, DCE-MRI breast cancer segmentation that is automated and accurate has great clinical application value and significance, while remaining a challenging task.
The present focus of mainstream research in automated segmentation of breast cancer is performed using deep learning strategies. In recent years, algorithms research based on deep learning has made great progress in medical image segmentation (Zhang et al. 2021; Pham et al. 2000). A mask-guided hierarchical fully convolutional network (FCN) segmentation framework was recently proposed at Duke University (Zhang et al. 2018). More specifically, a rough outline of the breast region and the accurate segmentation of breast cancer lesions was successfully performed using cascade FCN rough segmentation and FCN fine segmentation models. And their proposed models have been trained and tested on their own database, reaching a Dice coefficient of 0.72. However, troublesome issues are still encountered, such as missing, incorrect, or inaccurate segmentations of breast cancer lesions with small voxels. This is because only the FCN cascade architecture and a single-model structure were used. No effective solutions that might improve segmentation accuracy have been implemented, such as multi-scale and dynamic image data mechanisms. Such novel mechanisms are quite recent in development for accurate segmentation of breast cancer lesions.
Ronneberger et al. (2015) proposed a U-type network structure with encoder-decoder network architecture and a skip connection that has shown excellent performance in biomedical image segmentation. In the U-type structure, the image is continuously extracted and downsampled in the encoder. Briefly, different levels of feature information are extracted when reducing the image size to save memory resources. Such information is transmitted to the decoder through a skip connection. The decoder mines semantic information in the original image completely by combining high- and low-level feature information transmitted via the skip connection.
Semantic information in breast cancer segmentation tasks include the location, size, contour, boundary, and other characteristics of the lesion needed to perform automated segmentation. However, segmentation is vulnerable to noise, as breast cancer lesions are small while the larger area of the breast is complex with lobules, ducts, fatty and fibrous connective tissue, and so forth. Therefore, accuracy can be improved if normal breast tissue regions of interest can be delineated prior to segmentation (Zhang et al. 2018). Previous studies (Piantadosi et al. 2020; Ronneberger et al. 2015; Wei et al. 2018) have applied U-Net to separate the breast region from the breast wall line, demonstrating the ability and effectiveness of the U-Net network for breast ROI extraction.
The transformer structure proposed by Vaswani et al. (2017) has achieved good results in natural language processing (NLP) and other tasks (Han et al. 2021; Uddin et al. 2022), and researchers have been investigating its applicability for computerized visualization tasks (Li et al. 2020; Liu et al. 2021). For example, Dosovitskiy et al. (2020) proposed the Vision Transformer (ViT) structure for image classification tasks, which abandons the traditional Convolution Neural Network (CNN) structure completely and directly uses the Transformer encoder part to classify images. Furthermore, Carion et al. (2020) proposed the Detection with Transformer (DETR) structure for object detection tasks. This structure first uses CNN to extract features from the image and then inputs the feature sequence into the Transformer encoder to output the object category and coordinates. Prangemeier et al. (2020) added a multi-head attention branch and a decoder structure to DETR for cell instance segmentation. This structure employs DETR to detect cell instances while using the multi-head attention branch and the decoder structure to segment cell instances simultaneously. In addition, Zheng et al. (2020) proposed the Segmentation Transformer (SETR) structure for image semantic segmentation. This structure passes the image block directly into the Transformer encoder and upsamples the feature output from the Transformer through a decoder to perform the segmentation of different objects in the image.
Finally, Liu et al. (2021) have recently proposed a novel transformer (Swin Transformer) that could serve as the backbone of computer vision: a hierarchical transformer that can solve the differences caused when the visual entity scale is larger than the words used in the text. Medical image processing is an important field in computer visualization tasks, and we also believe that the Transformer structure has excellent performance characteristics. Therefore, the present study aims to achieve the precise segmentation of breast cancer lesions using Transformer structure.
Breast cancer lesions have small diameters that are variable in histological appearance, or blurry boundaries, whose surrounding background is complex. These aspects induce a typical class-imbalance problem. Tissues, such as muscle, heart, chest wall, skin, and fat, can seriously interfere with automated segmentation of breast cancer tumors. Considering that breast cancer lesions usually appear in the breast area, we propose a two-stage breast cancer segmentation framework to eliminate the influence of irrelevant factors as much as possible and obtain complete image information.
In the first stage, the U-Net model is trained to extract a rough breast ROI, eliminating the influence of breast muscle, fat, heart, and other irrelevant tissues.
Based on the breast ROI segmented in stage one, in the second stage, a TR-IMUnet model is proposed to perform breast tumor segmentation. The proposed model references the classic network U-Net. Then, the Transformer module, the improved dynamic rectified linear unit (IDy-ReLU) module, and the Multi-Scale Parallel Convolution Fusion (MSPCF) module were designed and integrated to construct the TR-IMUnet model. The model maximizes the advantages of the global and local receptive fields, obtains rich global information, extracts fine local information, and can dynamically adjust and retain data based on the differences in image data, thereby reducing image information loss and redundancy. The main work and contributions of this article are as follows:
-
(i)
Construction of the Transformer module (Vaswani et al. 2017) and integration into the U-Net network to maximize the advantages of the convolution operation and the Transformer module, conferring the model with a global receptive field and the ability to extract fine local information.
-
(ii)
Considering the shortcomings of the dynamic ReLU (Chen et al. 2020), an improved dynamic ReLU module is constructed, allowing the model to maximize the spatial and channel information of the input data to dynamically adjust and retain the data, reduce data loss and redundancy, and improve its robustness.
-
(iii)
In the coding path, an MSPCF module is proposed. A multi-scale parallel convolution structure is adopted, feature information of different scales is extracted through convolution operations of different kernel sizes, and the model’s ability to extract edge and fine information is enhanced.
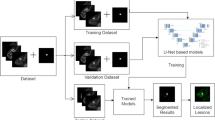
Experimental tests have shown that the proposed model performs well in breast cancer segmentation tasks, can accurately locate and segment breast cancer lesions in small areas, and has high segmentation accuracy (Fig. 1).
2 The proposed method
2.1 Breast ROI extraction
Breast cancer lesions do not generally appear at the chest wall or at the edge of the breast. Therefore, breast ROI extraction is conducive to the accurate segmentation of the tumor (Zhang et al. 2018). The benchmark U-Net model (Ronneberger et al. 2015) is commonly used in medical image segmentation tasks. In the first stage of the present study, the U-Net model was employed to achieve a rough breast ROI delineation based on the breast region labels outlined by clinicians (Fig. 2). Figure 2a, b shows a DCE-MRI breast cancer cross-sectional image. The ROI is marked in green, and the cancerous area is in yellow. A comparison of the two images demonstrates the variability of breast cancer tumors in shape, boundaries, cavities, and other features. The ITK-Snap tool was then used to construct a 3D rendering (Fig. 2c, d) of the spatial relationship between the breast tissue and the breast cancer.
2.2 Proposed TRIMU-Net construction method
A. Transformer model
Convolution is limited by the size of the convolution kernel, and the receptive field size is the same as the size of the convolution kernel. The traditional method increases the receptive field by stacking the convolutional and pooling layers together. However, this process makes the model deeper, and thus other issues are bound to emerge, such as gradient disappearance or gradient explosion, with an increased number of model parameters and calculations. Consequently, Vaswani et al. (2017) proposed the Transformer module, which was originally used for NLP tasks to obtain the dependency between each word. In the present study, the Transformer module was used for image segmentation to obtain the dependency between each pixel, i.e., the correlations information between long and short distances. Then, every pixel has a global receptive field. In contrast to Zheng et al. (2020), who only used the Transformer’s Encoder structure to extract global information, the present study used the U-Net encoder structure to extract features from the original image and pass them into the Transformer. This method compensates for the lack of local information caused by the loss of the Transformer’s ability to capture local features and maximizes the advantages of the convolution operation and the Transformer module. The specific structure of the applied Transformer module is shown in Fig. 3.
The Transformer module accepts the feature embedding of a 1D sequence \(Z\in {R}^{L\times C}\) as the input, where L is the sequence length and C is the hidden channel size. Therefore, the feature map needs to be converted from \(x\in {R}^{H\times W\times Z}\) to Z before passing into the Transformer. To save memory and reduce the number of calculations, only the Transformer was embedded into the last layer of the encoder structure. Conv-GN-ReLU was used to change the number of feature map channels, and thus meet the Transformer's needs. The specific conversion was:
A learnable position vector \(P \in R^{L \times C}\) as defined, with the same size as Z to encode the position information of each pixel. The Transformer input is \(E = Z + P\) The encoder structure of the Transformer module is composed of a multi-head self-attention and feedforward neural network. The multi-head self-attention has m layers, and each layer is a self-attention operation, which is used to obtain attention in different representation spaces. The self-attention formula is:
where \(Q = Z^{l} W_{Q} , K = Z^{l} W_{K} , V = Z^{l} W_{V}\) \(d_{k} = d_{model} = C/m\) and \(W_{Q} /W_{K} /W_{V} \in R^{C \times d}\) re the learnable parameters in the three linear transformation operations, and \(d\) is the dimension of\(Q/K/V\). Therefore, the formula for multi-head self-attention is as follows:
where \(W_{o} \in R^{md \times C}\) The feedforward network consists of two linear transformations and a ReLU activation function, and its formula is as follows:
where \( W_{1}\) nd \(W_{2}\) re the parameters of the two linear conversion layers and \(\sigma\) epresents the ReLU activation function.
Therefore, the output formula of the Transformer encoder part is
B. Improved dynamic ReLU activation function
The ReLU (Vinod et al. 2010) is widely used in deep neural networks because it can improve the performance of feedforward networks simply and effectively. The ReLU can be found in many typical structures, such as U-Net (Ronneberger et al. 2015), ResNet (He et al. 2015), and Unet3 + (Huang et al. 2020). ReLU-based improvements include leaky ReLU (Maas et al. 2013) and PReLU (He et al. 2015). However, according to Chen et al. (2020) most of these improvements (Maas et al. 2013; He et al. 2015) are static, i.e., they are performed in exactly the same way regardless of the input (such as images). Therefore, dynamic ReLU (Dy-ReLU), which uses data-adaptive approaches that automatically correct data according to different inputs, was proposed (Fig. 4). In the original article, the author described three different forms of Dy-ReLU, but only Dy-ReLU-B was deemed effective in our dataset, and therefore, the present study only discusses Dy-ReLU-B. For convenience, all subsequent Dy-ReLUs refer to Dy-ReLU-B. The main idea was to use a hyperfunction \(\theta (x)\) to encode the entire content of the input data and form an activation function \({f}_{\theta (x)}(x)\) that would be adaptive to the input data.
The definition of dynamic ReLU by Chen et al. (2020) is shown in formula (5):
where \(x_{c}\) epresents the data of the c-th channel of the input \(x\) and the coefficient \(({a}_{c}^{k},{b}_{c}^{k})\) is the output of the hyperfunction \(\theta (x)\):
where \(K\) s the number of functions and \(C\) s the number of channels. The parameters \({ }\left( {a_{c}^{k} ,b_{c}^{k} } \right){ }\) e related to all inputs x.
In Dy-ReLU (or Dy-ReLU-B), the hyperfunction \(\theta (x)\) is implemented by modeling the input data through a lightweight network similar to Squeeze-and-Excitation (SE) (Hu et al. 2018), as shown in the left part of Fig. 5. For an input vector x with \(C\times H\times W\) dimensions, the SE module first compresses the spatial information through global average pooling and then explicitly models the interdependence between the channels through two fully connected layers and a normalization layer, thereby recalibrating adaptively the channel characteristic response. Although the SE module is very effective, it still has shortcomings. For example, the squeeze operation of the SE module forces the output to lose its ability to capture interdependence among different spatial information. Dy-ReLU-C suggests that a branch should be built to supplement the spatial information. However, Dy-ReLU-C was experimentally shown to induce a negative effect. Thus, we referred to the Convolution Block Attention Module proposed by Woo et al. (2018) to improve the hyperfunction \(\theta (x)\), as shown in the right part of Fig. 5. The input vector \(x\) of \(C\times H\times W\) dimensions is compressed along the channel dimension through global average and global maximum pooling and then the convolutional and normalization layers are used to model the interdependence of spatial information explicitly. The spatial characteristic coefficients are multiplied by the original data to adaptively recalibrate the spatial characteristic response. At that point, the interdependence among different spatial information is captured, and the SE module captures the interdependence between the channels. The modified hyperfunction \(\theta \left( x \right) \) rmula is:
where \(M_{s}\) s the acquired spatial attention map and \(M_{s}\) s the acquired channel attention map. The definitions of \(M_{s}\) nd \(M_{c}\) re as follows:
where \(\sigma\) represents the \(Sigmoid\) activation function, \(\sigma^{\prime}\) represents the ReLU activation function, \({f}^{3\times 3}\) is the convolution operation with a 3 \(\times \) 3 convolution kernel, \(FFNN\) is the feedforward network, which is composed of two linear transformations and a ReLU function, \({W}_{1}\) and \({W}_{2}\) are the parameters of the two linear conversion layers, and \(Maxpool\) and \(Avgpool\) represent the global maximum pooling and global average pooling, respectively.
Similar to Dy-ReLU, \(2KC\) elements are the outputs that correspond to the coefficients \(a_{1:C}^{1:K}\) and \(b_{1:C}^{1:K}\) and are denoted as \(\Delta \alpha_{c}^{k} \left( x \right)\) and \(\Delta b_{c}^{k}\). The final output is calculated by adding the initial value and the output element in the following formula:
where \(\alpha^{k}\) and \(\beta^{k}\) are the initial values of \(\alpha_{c}^{k}\) and \(\beta_{c}^{k}\) respectively, and \(\lambda_{a}\) and \(\lambda_{b}\) re scalars that control the coefficient range. In accordance with Chen et al. (2020), we let \(K = 2\) and set \(\alpha^{1} = 1,\alpha^{2} = \beta^{1} = \beta^{2} = 0\) \(\lambda_{a}\) and \(\lambda_{b}\) default to 1.0 and 0.5, respectively.
C. MSPCF module
The MSPCF module was introduced because cancerous areas vary in size, shape, gray scale, and definitive boundary (Fig. 6). With the MSPCF module parallel convolution structures can obtain the characteristics of different receptive fields in the same layer, improve the accuracy of segmentation in cases of blurred edges and small masses, and then transfer the information of different scales to the next layer. Consequently, this can provide rich semantic information for the subsequent down- and up-sampling of the decoder layer, thereby overall improving the segmentation accuracy.
The MSPCF module contains three parallel convolutional layers and one global average pooling layer. These parallel convolution operations have \(1\times 1\), \(3\times 3,\) and \(5\times 5\) kernel sizes. The multi-scale context feature information is obtained by using the convolution of different kernel sizes, and the \(1\times 1\) convolution layer is used to retain the feature information of the current scale. The MSPCF module also uses the global average pooling layer to obtain pixel-level global information, and then use bilinear interpolation to obtain the required dimensions. Using the \(1\times 1\) convolution process, the global average pooling layer information is fused and compressed with the three parallel convolutional layers. Finally, the output feature map is passed to the next layer.
D. Improvement process
The Transformer improved dynamic ReLU, and multi-scale parallel convolution modules were added to the benchmark U-Net model to discuss the effectiveness of each module. Finally, all modules were added to the benchmark U-Net model to form the proposed TR-IMUnet model that achieved the best performance. Improvement was in accordance with the following protocol:
First, based on the classic U-Net network, the Transformer module was introduced to capture the global information of the image and form a global attention mechanism so that each pixel had a global receptive field. To allow feature extraction, and thus compensate for the feature loss caused by the Transformer’s inability to capture local information, the encoder of the U-Net model was used. Thus, the new model should maximize the convolution operation and the Transformer module. This model was named TR-U-Net.
Second, based on the dynamic ReLU proposed by Chen et al. (2020), we verified the effectiveness of the TR-U-Net model based on the benchmark U-Net model and identified its potential shortcomings experientially. Thereby, an improved dynamic ReLU module was designed to replace the ReLU operation in the U-Net model. Each activation function in the model was adaptive and specifically designed according to the input spatial data and channel information, thus maximizing the input information and improving the generalization ability of the model. This model was called Improved U-Net (IUnet). The Transformer module and the improved dynamic ReLU module were added into the benchmark U-Net model to combine the advantages of each module and form the TR-IUnet model.
Finally, based on the proposed TR-IUnet network, the MSPCF module was embedded behind the feature extraction unit of each layer of the coding part, which was used to extract the feature information of different levels and scales, and then transmit it to the next layer after fusion and compression to enrich the feature information extracted by the network. At the same time, this is transmitted to the corresponding decoder layer through a long connection to supplement the semantic information lost during the upsampling process. Experimental tests showed that the TR-IMUnet model can enable optimal breast cancer segmentation.
E. Loss function
Data imbalance is a common issue in medical image segmentation. In most cases, the number of lesion voxels in the dataset is significantly less than the number of non-lesion voxels. The same is true for breast cancer, and the area of lesions is much smaller than that of the breast area. For this purpose, the Tversky (Salehi et al. 2017) loss function was adopted. Based on the Tversky index, this loss function can effectively solve the problem of data imbalance, allowing for a good balance between precision and recall rate. The formula is as follows:
where \(p,g\) are the output result of the last \(sigmoid\) layer of the network. Among them, \(p_{0i}\) is the output probability of the pathological voxel, and \(p_{1i}\) is the output probability of the non-pathological voxel. Similarly, \(g\)\(g_{0i}\) represent the output probability of a pathological voxel, and \(g_{1i}\) represents the output probability of a non-pathological voxel. Then, \(\alpha ,\beta\) control the false negatives and false positives separately. We can control the trade-off between false positives and false negatives by adjusting \(\alpha ,\beta\). In our experimental process, we set \(\alpha = 0. 3 {\text{and}} \beta = 0.7\)
3 Experimental process and analysis
3.1 Experimental environment
This study used an Intel Core I7-8700 K CPU with 3.20 GHz base frequency as server hardware equipped with 32 GB of memory. We also used a GeForce RTX graphics card, Win10 operating system, and Python 3.6 as the programming language. A deep learning framework based on Pytorch1.3 was used.
3.2 Dataset and preprocessing
The DCE-MRI imaging data used in this experiment originated from a self-built clinical database consisting of 160 cases. A Siemens MAGNETO 1.5 T MRA was used for the acquisition of breast cancer images, equipped with a special four-channel phased array surface coil. All subjects were in the prone position during the acquisition process. After screening, clinical data from 160 cases of DCE-MRI stage T2 were finally included in the database. Images were obtained in the transverse direction, and the image size was 512 × 512. All breast ROIs were manually marked by the clinician with the LabelMe software, whereas 3D slicer was used to label the breast cancer cells. The 160 cases were randomly divided into 128 cases in the training set and 32 cases in the test set. We then performed data enhancement operations, such as mirroring, scaling, and elastic deformation, during the training process. In the training process, the data were effectively preprocessed. Initially, the first 0.1% and the last 0.1% of pixel values were deleted for denoising, and then gray scale normalization was performed. For breast ROI region extraction and breast cancer lesion segmentation, the same batch of training and test sets were used. During the training process, both models were trained successively. First, we used the U-Net model for breast ROI extraction. Second, we used the \(He\) normalization algorithm to initialize the breast cancer segmentation model during the training process. Adam Optimizer (Kingma and Ba 2014) performed gradient descent, with the parameters set to \({\beta }_{1}=0.9, {\beta }_{2}=0.999, \varepsilon ={le}^{-8}\), and the learning rate was \({le}^{-8}\). The batch size was set to 3, and Early Stopping (Prechelt 1998) was used to monitor the training process. Finally, the output result was obtained through the \(sigmoid\) function. Because the range of the output value was (0,1), the final output result was assessed by setting the threshold to 0.5.
3.3 Segmentation index
Five commonly used medical image segmentation indexes were used to evaluate the segmentation results: Dice coefficient (Dice), Intersection over Union (IoU), Sensitivity (SEN), Specificity (SPE), and Positive predictive value (PPV), and the formula is shown in (5). TP is the number of pixels with the correct foreground segmentation in pixel-level segmentation, TN is the number of pixels with the correct background segmentation in pixel-level segmentation, FP is the number of pixels with background segmentation error in pixel-level segmentation, and FN is the foreground segmentation error in pixel-level segmentation.
3.4 Breast ROI segmentation
The benchmark U-Net model was used to roughly delineate the breast ROI, and the respective segmentation diagram is shown in Fig. 7. The first row depicts images from three patients with breast cancer. The second row shows how the breast region labels were manually segmented by the clinician. Finally, the red area in the third row demonstrates the rough segmentation result of the breast ROI obtained by the U-Net model. The breast area can be extended to the sides of the chest to ensure that no cancerous area is missed. Therefore, even small breast ROIs could be segmented correctly.
The experimental statistics of the segmentation results showed that the Dice and ACC values could reach 0.92 and 0.98, respectively, as shown in Table 1. These values meet the requirement for the precise segmentation of the breast cancer area described in the next step. It should be mentioned that the segmentation of breast cancer lesions remained unaffected.
3.5 Experimental comparative analysis
Using the same limited dataset, the proposed model was then compared with current mainstream segmentation models with excellent performance. Our improved models were TR-U-Net, IUnet, TR-IUnet, and TR-IMUnet. Five excellent mainstream medical image segmentation models were selected for a comparative analysis: U-Net (Ronneberger et al. 2015), 2D-VNet (Milletari et al. 2016), ResUnet (He et al. 2016), 2D-DenseUnet (Li et al. 2018), and Attention-U-Net (Oktay et al. 2018) (Figs. 8, 9, and 10).
Figure 8 illustrates the segmentation results of three patients with breast cancer lesions using the selected five mainstream segmentation models. The first row depicts the original images obtained from the three patients, the second row represents their corresponding labels, and rows 3–7 correspond to the segmentation results of the U-Net, 2D-VNet, ResUnet, 2D Dense-Unet, and Attention-U-Net models, respectively. It is shown that the U-Net segmentation result in the third row had rough edges, and thus it was impossible to distinguish the normal gland from the breast cancer lesion area. Similarly, the ResUnet segmentation result shown in the fifth row underlines that the lesion could not be distinguished from the surrounding normal glands. Hence, the segmentation result can often be smaller than the label area. The fourth row demonstrates the 2D-VNet segmentation result. In this case, the lesions could not be segmented due to the obstructive effect of the breast edge and surrounding glands. Furthermore, the sixth row shows the 2D Dense-UNet segmentation result. Problems of over-segmentation or under-segmentation emerged when segmenting this model because the lesion boundary had blurry characteristics. Finally, the seventh row depicts the Attention-Unet segmentation result. From a holistic point of view, the model uses the attention mechanism to locate the lesion accurately; however, edges and details are still lacking.
In contrast, the segmentation results of the four improved models proposed in this paper are shown in rows 4–7 of Fig. 9. The third row depicts the segmentation effect of Dy-ReLUBUnet, which incorporated the Dy-ReLU-B module proposed by Chen et al. (2020) into the U-Net. The segmentation effect was slightly improved after fusing the Dy-ReLU-B module, which can basically segment the contour of the lesion, but still failed to provide fine details. The fourth row shows the segmentation effect of the improved Dy-ReLU-B model, i.e., IUnet. This model could distinguish the lesion edge more finely, and the segmentation area was significantly closer to the label area. The fifth row is the segmentation result of the TR-Unet, which incorporated the Transformer module into U-Net. This model could capture the global information of the image markedly better than the previous models, and could thus localize the lesions accurately. The sixth row, which represents the TR-IUnet model, highlights that this model offers the same advantages with the improved Dy-ReLU but it can also capture the global information of the image, thereby providing useful information in breast cancer segmentation and refining the segmentation results. Finally, the last row reflects the TR-IMUnet model, which exhibits the highest segmentation accuracy among all experimental networks. This model adds a multi-scale fusion mechanism on the basis of TR-IUnet and provides semantic information for segmentation tasks by fusing information from different scales, thereby improving the segmentation effect.
Figure 10 uses a single patient case to analyze the segmentation results from a global perspective. Comparison between Fig. 10 (d) and (g) reveals that the Transformer characteristics are similar to the attention structure, and they both contribute to the accurate localization of the lesion. The difference is that the attention structure is local attention with a small receptive field, whereas Transformer has a global receptive field. The improved dynamic ReLU module focuses on the shortcomings of the existing module, and it successfully maximizes not only the spatial context information, but also the dependencies between channels. Therefore, lesions are segmented more precisely. Meanwhile, the proposed TR-IMUnet model, which has the advantage of integrating the Transformer module while improving dynamic ReLU, can efficiently add multi-scale modules and provide semantic information under different scales and representations for model learning, thereby improving model performance and segmentation accuracy. Compared with all the experimental networks in this study, our proposed model shows the highest segmentation accuracy, suggesting that it has better robustness and compatibility.
3.6 Module feature analysis
A. Global receptive field of the Transformer module
In the second section, it was mentioned that the greatest advantage of the Transformer module is that all pixels have a global receptive field, which allows the model to capture global information and model the relationship between pixels in an explicit manner. Figure 11 highlights this feature. We initially selected a random point and displayed it as an image. Each pixel could roughly capture the original image information, which is represented as the outline of the breast region in the breast cancer data set. In addition, the color distribution demonstrates that different pixels provide different image information. For example, lesion pixels have a strong connection with the lesion and a weak connection with the background and other glands, whereas background pixels have a strong connection with the background and a weak connection with other pixels. Moreover, other gland pixels have a strong connection with the lesion and other glands, which is consistent with our cognition. (Note that the yellow color reflects a strong correlation as opposed to the blue color which reflects a weak correlation.) In other words, the Transformer module can make all pixels have global receptive fields, and can also explicitly model the interdependence between each pixel. This is very practical and helpful for lesion identification and fine segmentation.
B. Multi-scale function of the MSPCF module
The primary goal of medical image segmentation is to obtain a binary image that contains the lesion location only (lesion location is 1, and the rest is 0). Therefore, our neural network model should have the ability to identify and highlight lesion location and weak non-lesion location. The output features of each layer of the network in Fig. 12 show that the continuous feature extraction of the CNN on the input image through continuous convolution operation is indeed a process of constantly highlighting the lesion and weakening the non-lesion parts. This meets our needs. However, a single convolution operation cannot clearly distinguish between lesion and non-lesion areas, as shown in Fig. 12(c). Some noise on the edge may be misdiagnosed as lesion areas. The MSPCF module can correct or confirm the identification of the lesion and non-lesion areas in the image by fusing feature information of different scales to reduce the probability of misjudgment, as shown in Fig. 12g. Comparison of Fig. 12d and i, as well as Fig. 12e and j, reveals that the MSPCF module can further highlight the lesion location.
C. Dynamic performance of the improved dynamic ReLU module
In the second section, it was stated that the greatest advantage of dynamic ReLU is its ability to construct a specific activation function in accordance with different input data, and to dynamically retain and adjust these input data to reduce data loss or redundancy. As shown in Fig. 13, the ReLU activation functions present different distributions for different stages, and the activation function of each channel in each stage is also different. In addition, the slope of each ReLU activation function is also different. For example, the slope of the positive axis of ReLU in blocks 5 and 6 is approximately one, whereas the slope of the positive axis of ReLU in blocks 2 to 4 is approximately two, indicating that dynamic ReLU can filter the input data, and also enlarge or reduce these data according to their importance.
Consequently, this renders the model more flexible and with enhanced anti-interference ability.
D. Model visualization
Finally, the entire process of breast cancer segmentation was reviewed by visualizing each stage of the proposed model. When a breast cancer image is input into the TR-IMUnet model, the encoder of the TR-IMUnet identifies and highlights the lesion, and subsequently weakens the non-lesion area via continuous convolution layers (Fig. 14). At the same time, the multi-scale layer can further correct or confirm the location of the lesion by fusing feature information of different scales. The Transformer layer can compensate for the narrow receptive field of the convolution layer by exploiting its global receptive field, maximizing the global and local feature information, and transferring the acquired features to the decoder layer. Consequently, the decoder layer receives high-level and low-level feature information from the encoder layer. On the basis of the original image space information provided by the low-level feature information, the abstract feature information is restored to the corresponding position of the original image and is separated from the surrounding glands. Thus, we can obtain high-brightness and grayscale images that represent lesions and non-lesions, respectively. Finally, after the thresholding operation and the activation of the sigmoid function, a segmented image containing only the lesion is obtained.
3.7 Stereoscopic segmentation results
For the effective diagnosis of breast cancer, the position or area on the 2D image, but also the shape, volume, and size of the entire cancer must be considered. This information is vital for the preoperative preparation and postoperative rehabilitation of breast cancer resection. Therefore, the 2D segmentation image of the model was transformed into a 3D structure to illustrate the segmentation results. Figure 15 shows that the present proposed model can efficiently segment the 3D outline of the entire breast lesion, and its outline size is basically consistent with the outline size marked by professional radiologists. The proposed model can also correctly segment breast lesions with different shapes. For instance, the first three rows (Fig. 15) depict hollow, solid, and scattered blocks, respectively. Furthermore, the proposed TR-IMUnet model can restore the 3D structure of the breast lesions and achieve the highest segmentation accuracy by maximizing the advantages of each component module.
3.8 Statistical analysis
To verify the segmentation performance of the proposed model for breast lesions, the evaluation indexes Dice, IoU, Sensitivity, Specificity, and PPV were, respectively, applied to the dataset, and the results were analyzed statistically. It should be mentioned that all models in the table are based on the two-stage segmentation framework. First, the U-Net model was used to extract the breast ROI. Then different models were employed to segment breast cancer.
The segmentation accuracies of the ResUnet, DenseUnet, Att-U-Net, DyReluB-U-net, TR-U-Net, IUnet, TR-IUnet, and TR-IMUnet models were each better than the benchmark U-Net model (Table 2). The segmentation accuracy of DyReluBUnet was slightly better than that of the benchmark U-Net, as evidenced by the increase in Dice by 2.11%. The IUnet model with an improved dynamic ReLU module improved the segmentation accuracy by 1.08 while the performance of the TR-U-net with the Transformer module was similar. Finally, TR-IMUnet, which combines the advantages of both Transformer and Improved Dynamic ReLU and adds the MSPCF module, achieved the highest segmentation accuracy.
Compared with the benchmark U-Net, the Dice coefficient of the TR-IMUnet model is increased by 4.27%, IoU increases by 5.21%, sensitivity increases by 3.37%, and PPV increases by 3.68%. A segmentation comparison diagram of U-Net and TR-IMUnetis shown in Fig. 16. As shown on the left side of Fig. 16, when the U-Net model is used to segment 2D tumor images, there are significant problems, such as improper detection of the shape, size, and contour of breast tumors, resulting in missing, incorrect or even large-area loss of segmentation results. Moreover, the second column of the third row in Fig. 16a demonstrates that U-Net performs poorly when accurate segmentation is used in hard samples because it cannot distinguish between lesion and the non-lesion positions. In contrast, the proposed TR-IMUnet model can accurately locate the position of breast tumors, detect the size and shape of tumor blocks correctly, and distinguish them from non-lesion areas far more efficiently. At the same time, when detecting difficult images, the proposed model can also maintain high segmentation accuracy and segment the tumor meticulously. Moreover, by observing the segmentation results of U-Net and TR-IMUnet models from the 3D perspective of Fig. 16b, it can be found that the U-Net segmentation model performance is ordinary, and the performance of the proposed TR-IMUnet model is better than that of U-Net in fine segmentation of the size, shape and details of breast tumor.
Figure 17 provides a statistical comparison of various algorithm BOX charts. The horizontal axis represents different models used in previous experimental comparison, whereas the vertical axis represents the Dice coefficients. After the improvement process, the index values and stability of the proposed IUnet, TR-U-net, TR-IUnet, and TR-IMUnet models gradually increased, verifying the effectiveness of the proposed modules and the validity of the proposed model improvement.
3.9 Analysis of model parameters
The parameters used in the proposed model were approximately 13.9 MB (Table 3). The proposed model replaces ReLU in the benchmark U-Net encoder-decoder structure based on the proposed improved dynamic ReLU module, i.e., Conv → GroupNorm → Improved Dy-ReLU → Conv → GroupNorm → Improved Dy-ReLU.
The MSPCF module is added to the coding path of TR-IMUnet, which contains four encode layers and four maximum pooling operations, and the area of the feature graph is halved after each operation. Except for MSPCF, all convolution layers have 3 × 3 convolution kernels, and the number of channels is shown in the above table. The first encoding layer outputs 32 channels. With the deepening of the network, the number of channels is doubled after each coding layer and the maximum pooling operation. The number of channels remains at 256 up until the last coding layer, and then the channels are compressed to 128, and the feature information will be transmitted to the Transformer.
In the decoder part, after upsampling the feature map, the decoder still adopts the same operation as the encoder infrastructure, i.e., Conv → GroupNorm → Improved Dy-ReLU → Conv → GroupNorm → Improved Dy-ReLU. After each operation, the area of the feature map is twice as large as the original, and the channel parameters decrease by half. The output of the upsampling operation is connected to the output of the corresponding part of the encoder as the input of the lower layer. The resulting feature map is processed by convolution to maintain the same number of channels as that of the symmetric encoder part. Finally, the final result is output by one-time 1 × 1 convolution.
3.9.1 Training and validation
The training loss of the present proposed model converged rapidly between none and 35 times (Fig. 18). In the subsequent iterations, the loss function curve gradually converged and tended to flatten, and the convergence speed of the verification loss curve was faster between none and 25 iterations. The curve convergence tended to be flat in subsequent iterations. The early stop method was triggered to stop the training process when the loss remained constant for a certain number of rounds, to save time and resources. In summary, in training the proposed model was robust.
4 Conclusions
This study introduced a two-stage breast cancer image segmentation model. The U-Net model was used first to obtain a rough outline of the breast region. Then, a TR-IMUnet model was designed, in which the ReLU function of the encoder-decoder structural unit was replaced by an improved dynamic ReLU function, and the MSPCF and Transformer modules were added at the encoder path and end, respectively. Compared with the U-Net benchmark model, the Dice, IoU, SEN, and PPV breast cancer segmentation indexes were improved by 4.27, 5.21, 3.37, and 3.68%, respectively. The segmentation of small mesh lesions warrants further improvement. To improve the diagnosis, treatment, and prognosis of breast cancer, in the future we intend to study the 3D segmentation and location of breast cancer images and establish a prediction model in combination with Radiomics.
Data availability
The datasets and the code generated and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Carion N, Massa F, Synnaeve G, et al (2020) End-to-end object detection with transformers. In: European conference on computer vision. Springer, Cham, pp 213–229
Chen Y, Dai X, Liu M, et al (2020) Dynamic relu. In: European conference on computer vision. Springer, Cham, pp 351–367
Dosovitskiy A, Beyer L, Kolesnikov A, et al (2020) An image is worth 16x16 words: transformers for image recognition at scale. arXiv preprint arXiv:2010.11929
Han K, Xiao A, Wu E, et al (2021) Transformer in transformer. Adv Neural Inf Process Syst 34
He K, Zhang X, Ren S, et al (2015) Delving deep into rectifiers: Surpassing human-level performance on imageNet classification. In: Proceedings of the IEEE international conference on computer vision, pp 1026–1034
He K, Zhang X, Ren S, et al (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Hu J, Shen L, Sun G (2014) Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7132–7141
Huang H, Lin L, Tong R, et al (2020) Unet 3+: A full-scale connected unet for medical image segmentation. In: ICASSP 2020–2020 IEEE international conference on acoustics, speech and signal processing (ICASSP). IEEE, pp 1055–1059
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980
Li X, Chen H, Qi X et al (2018) H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans Med Imaging 37(12):2663–2674
Li Z, Liu X, Creighton FX, et al (2020) Revisiting stereo depth estimation from a sequence-to-sequence perspective with transformers. arXiv preprint arXiv:2011.02910
Litjens G, Kooi T, Bejnordi BE et al (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88
Liu R, Yuan Z, Liu T, et al (2021) End-to-end Lane shape prediction with transformers. In: Proceedings of the IEEE/CVF winter conference on applications of computer vision, pp 3694–3702.
Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, Lin S, Guo B (2021) Swin transformer: Hierarchical vision transformer using shifted windows. arXiv preprint arXiv:2103.14030
Maas AL, Hannun AY, Ng Y (2013) Rectifier nonlinearities improve neural network acoustic models. Proc. Icml. 30(1): 3.
Milletari F, Navab N, Ahmadi SA (2016) V-net: Fully convolutional neural networks for volumetric medical image segmentation[C]//2016 fourth international conference on 3D vision (3DV). IEEE, 2016: 565–571.
Nair V, Hinton GE (2010a) Rectified linear units improve restricted boltzmann machines
Nair V, Hinton GE (2010b) Rectified Linear Units Improve Restricted Boltzmann Machines. International Conference on Machine Learning
Oktay O, Schlemper J, Folgoc L L, et al (2018) Attention u-net: Learning where to look for the pancreas. arXiv preprint arXiv:1804.03999
Pham DL, Xu C, Prince JL (2000) Current methods in medical image segmentation[J]. Annu Rev Biomed Eng 2(1):315–337
Piantadosi G, Sansone M, Fusco R et al (2020) Multi-planar 3D breast segmentation in MRI via deep convolutional neural networks. Artif Intell Med 103:101781
Prangemeier T, Reich C, Koeppl H (2020) attention-based transformers for instance segmentation of cells in microstructures. In: 2020 IEEE international conference on bioinformatics and biomedicine (BIBM). IEEE, pp 700–707
Prechelt L (1998) Early stopping-but when? [M]//Neural Networks: Tricks of the trade. Springer, Berlin, Heidelberg, pp 55–69
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 234–241
Salehi SSM, Erdogmus D, Gholipour A (2017) Tversky loss function for image segmentation using 3D fully convolutional deep networks[C]//International workshop on machine learning in medical imaging. Springer, Cham, pp 379–387
Uddin MN, Li B, Ali Z et al (2022) Software defect prediction employing BiLSTM and BERT-based semantic feature. Soft Comput. https://doi.org/10.1007/s00500-022-06830-5
Vaswani A, Shazeer N, Parmar N, et al (2017) Attention is all you need. arXiv preprint arXiv:1706.03762
Wei D, Weinstein S, Hsieh MK et al (2018) Three-dimensional whole breast segmentation in sagittal and axial breast MRI with dense depth field modeling and localized self-adaptation for chest-wall line detection. IEEE Trans Biomed Eng 66(6):1567–1579
Woo S, Park J, Lee J Y, et al (2018) Cbam: convolutional block attention module[C]//Proceedings of the European conference on computer vision (ECCV), pp 3–19
Xiao J, Rahbar H, Hippe DS et al (2021) Dynamic contrast-enhanced breast MRI features correlate with invasive breast cancer angiogenesis. NPJ Breast Cancer 7:42
Zhang J, Saha A, Zhu Z et al (2018) Hierarchical convolutional neural networks for segmentation of breast tumors in MRI with application to radiogenomics. IEEE Trans Med Imaging 38(2):435–447
Zhang K, Shi Y, Hu C et al (2021) Nucleus image segmentation method based on GAN and FCN model. Soft Comput. https://doi.org/10.1007/s00500-021-06449-y
Zheng S, Lu J, Zhao H, et al (2020) Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. arXiv preprint arXiv:2012.15840
Funding
This study is supported by the National Nature Science Foundation of China (No.62071213), Special Project in key Areas of Artificial Intelligence in Guangdong Universities (No.2019KZDZX1017), Guangdong Basic and Applied Basic Research Foundation (No. 2019A1515010716), and open fund of Guangdong Key Laboratory of digital signal and image processing technology (2019GDDSIPL-03, 2020GDDSIPL-03).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Qin, C., Wu, Y., Zeng, J. et al. Joint Transformer and Multi-scale CNN for DCE-MRI Breast Cancer Segmentation. Soft Comput 26, 8317–8334 (2022). https://doi.org/10.1007/s00500-022-07235-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-022-07235-0