Abstract
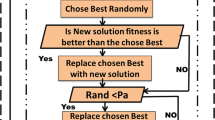
A modified Artificial Bee Colony (ABC) metaheuristics optimization technique is applied for cancer classification, that reduces the classifier’s prediction errors and allows for faster convergence by selecting informative genes. Cuckoo search (CS) algorithm was used in the onlooker bee phase (exploitation phase) of ABC to boost performance by maintaining the balance between exploration and exploitation of ABC. Tuned the modified ABC algorithm by using Naïve Bayes (NB) classifiers to improve the further accuracy of the model. Independent Component Analysis (ICA) is used for dimensionality reduction. In the first step, the reduced dataset is optimized by using Modified ABC and after that, in the second step, the optimized dataset is used to train the NB classifier. Extensive experiments were performed for comprehensive comparative analysis of the proposed algorithm with well-known metaheuristic algorithms, namely Genetic Algorithm (GA) when used with the same framework for the classification of six high-dimensional cancer datasets. The comparison results showed that the proposed model with the CS algorithm achieves the highest performance as maximum classification accuracy with less count of selected genes. This shows the effectiveness of the proposed algorithm which is validated using ANOVA for cancer classification.









Similar content being viewed by others
Data availability
All used data are benchmark high-dimensional microarray datasets of cancer and are freely available in different repositories.
References
Alomari OA, et al. (2021) Gene selection for microarray data classification based on Gray Wolf Optimizer enhanced with TRIZ-inspired operators. Knowl Based Syst 223: 107034.
Alon U et al (1999) Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci 96(12):6745–6750
Alshamlan HM, Badr GH, Alohali YA (2015) Genetic bee colony (GBC) algorithm: a new gene selection method for microarray cancer classification. Comput Biol Chem 56:49–60
Armstrong SA et al (2002) MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet 30(1):41–47
Aziz R, Verma C, Srivastava N (2016) A fuzzy based feature selection from independent component subspace for machine learning classification of microarray data. Genom Data.
Aziz R, Verma C, Srivastava N (2017a) Dimension reduction methods for microarray data: a review. AIMS Bioeng 4(2):179–197
Aziz R et al (2017b) Artificial neural network classification of microarray data using new hybrid gene selection method. Int J Data Min Bioinform 17(1):42–65
Aziz R, Verma C, Srivastava N (2017c) A novel approach for dimension reduction of microarray. Comput Biol Chem.
Aziz RM, Hussain A, Sharma P, Kumar P (2022a) Machine learning-based soft computing regression analysis approach for crime data prediction. Karb Int J Mod Sci 8(1):1–19
Aziz RM, Baluch MF, Patel S, Ganie AH (2022b) LGBM: a machine learning approach for Ethereum fraud detection. Int J Inf Technol 13(1):1–11
Baburaj E (2022) Comparative analysis of bio-inspired optimization algorithms in neural network-based data mining classification. Int J Swarm Intell Res (IJSIR) 13(1):1–25
Chen X, Yu K (2019) Hybridizing cuckoo search algorithm with biogeography-based optimization for estimating photovoltaic model parameters. Sol Energy 180:192–206
Coleto-Alcudia V, Vega-Rodríguez MA (2020) Artificial Bee Colony algorithm based on Dominance (ABCD) for a hybrid gene selection method. Knowl Based Syst 205:106323
Cristin R et al (2020) Deep neural network based rider-cuckoo search algorithm for plant disease detection. Artif Intell Rev 2020:1–26
Cui Z et al (2019) A hybrid many-objective cuckoo search algorithm. Soft Comput 23(21):10681–10697
Dash R (2021) An adaptive harmony search approach for gene selection and classification of high dimensional medical data. J King Saud Univ Comput Inform Sci 33(2):195–207
De Campos LM, et al. (2011) Bayesian networks classifiers for gene-expression data. In: Intelligent Systems Design and Applications (ISDA), 2011 11th International Conference on 2011. IEEE.
Desai NP et al (2022) Image processing model with deep learning approach for fish species classification. Turk J Comput Math Educ 13(1):85–99
Ding Z, Lu Z, Liu J (2018) Parameters identification of chaotic systems based on artificial bee colony algorithm combined with cuckoo search strategy. Sci China Technol Sci 61(3):417–426
Dwivedi AK (2018) Artificial neural network model for effective cancer classification using microarray gene expression data. Neural Comput Appl 29(12):1545–1554
Elek J, Park K, Narayanan R (1999) Microarray-based expression profiling in prostate tumors. In Vivo (Athens Greece) 14(1):173–182
Fan L, Poh K-L, Zhou PJESWA (2009a) A sequential feature extraction approach for naïve bayes classification of microarray data 36(6): 9919–9923
Fan L, Poh K-L, Zhou P (2009b) A sequential feature extraction approach for naïve bayes classification of microarray data. Expert Syst Appl 36(6):9919–9923
Friedman N, Geiger D, Goldszmidt M (1997) Bayesian network classifiers. Mach Learn 29(2):131–163
Garro BA, Rodríguez K, Vázquez RA (2015) Classification of DNA microarrays using artificial neural networks and ABC algorithm. Appl Soft Comput.
Garro BA, Rodríguez K, Vázquez RA (2016) Classification of DNA microarrays using artificial neural networks and ABC algorithm. Appl Soft Comput 38:548–560
Golub TR et al (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286(5439):531–537
Gordon GJ et al (2002) Translation of microarray data into clinically relevant cancer diagnostic tests using gene expression ratios in lung cancer and mesothelioma. Can Res 62(17):4963–4967
Hall M (2007) A decision tree-based attribute weighting filter for naive Bayes. Knowl Based Syst 20(2):120–126
Hameed SS et al (2021) A comparative study of nature-inspired metaheuristic algorithms using a three-phase hybrid approach for gene selection and classification in high-dimensional cancer datasets. Soft Comput 2021:1–19
Hasan BMS, Abdulazeez AM (2021) A review of principal component analysis algorithm for dimensionality reduction. J Soft Comput Data Mining 2(1):20–30
Hsu C-C, Chen M-C, Chen L-S (2010) Integrating independent component analysis and support vector machine for multivariate process monitoring. Comput Ind Eng 59(1):145–156
Hyvarinen A, Karhunen J, Oja E (2001) Independent component analysis. Wiley
Jatoth RK, Rajasekhar A (2010) Speed control of pmsm by hybrid genetic artificial bee colony algorithm. In: Communication Control and Computing Technologies (ICCCCT), 2010 IEEE International Conference on IEEE
Kıran MS et al (2012) A novel hybrid approach based on particle swarm optimization and ant colony algorithm to forecast energy demand of Turkey. Energy Convers Manage 53(1):75–83
Kumar L, Bharti KKJNC (2021) A novel hybrid BPSO–SCA approach for feature selection. Natl Comput 20(1): 39–61.
Li G et al (2017) Prediction of biomarkers of oral squamous cell carcinoma using microarray technology. Sci Rep 7:42105
Li J et al (2021) Multi-source feature extraction of rolling bearing compression measurement signal based on independent component analysis. Measurement 172:108908
Lv J et al (2016) A multi-objective heuristic algorithm for gene expression microarray data classification. Expert Syst Appl 59:13–19
Mafarja M et al (2020) Efficient hybrid nature-inspired binary optimizers for feature selection. Cogn Comput 12(1):150–175
Mahdavi K, Labarta J, Gimenez J (2019) Unsupervised feature selection for noisy data. In: International Conference on Advanced Data Mining and Applications. Springer.
Mollaee M, Moattar MH (2016) A novel feature extraction approach based on ensemble feature selection and modified discriminant independent component analysis for microarray data classification. Biocybern Biomed Eng 36(3):521–529
Mollaee M, Moattar MHJB, Engineering B (2016) A novel feature extraction approach based on ensemble feature selection and modified discriminant independent component analysis for microarray data classification. Biocybern Biomed Eng 36(3):521–529
Musheer RA, Verma CK, Srivastava N (2019) Novel machine learning approach for classification of high-dimensional microarray data. Soft Comput 23(24):13409–13421
Nutt CL et al (2003) Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Can Res 63(7):1602–1607
Ong HF, et al (2020) Informative top-k class associative rule for cancer biomarker discovery on microarray data 146: 113169.
Othman MS, Kumaran SR, Yusuf LM (2020) Gene selection using hybrid multi-objective cuckoo search algorithm with evolutionary operators for cancer microarray data. IEEE Access 8:186348–186361
Pandey AC, Rajpoot DS (2019) Spam review detection using spiral cuckoo search clustering method. Evol Intel 12(2):147–164
Pandey AC, Rajpoot DS, Saraswat M (2020) Feature selection method based on hybrid data transformation and binary binomial cuckoo search. J Ambient Intell Humaniz Comput 11(2):719–738
Peng H et al (2021) Multi-strategy serial cuckoo search algorithm for global optimization. Knowl Based Syst 214:106729
Rabia A, Namita S, Chandan KV (2015) A weighted-SNR feature selection from independent component subspace for NB classification of microarray data. Int J Adv Biotechnol Res 6(2):245–255
Salem H, Attiya G, El-Fishawy N (2017) Classification of human cancer diseases by gene expression profiles. Appl Soft Comput 50:124–134
Selaru F et al (2002) Global gene expression profiling in Barrett’s esophagus and esophageal cancer: a comparative analysis using cDNA microarrays. Oncogene 21(3):475–478
Shehab M, Khader AT, Al-Betar MA (2017) A survey on applications and variants of the cuckoo search algorithm. Appl Soft Comput 61:1041–1059
Singh D et al (2002) Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1(2):203–209
Song P-C, Pan J-S, Chu S-C (2020) A parallel compact cuckoo search algorithm for three-dimensional path planning. Appl Soft Comput 94:106443
Turgut S, Dağtekin M, Ensari T (2018) Microarray breast cancer data classification using machine learning methods. In: 2018 Electric Electronics, Computer Science, Biomedical Engineerings' Meeting (EBBT). IEEE.
Venkatesh B, Anuradha J (2019) A review of feature selection and its methods. Cybern Inform Technol 19(1):3–26
Wang X-H et al (2020) Multi-objective feature selection based on artificial bee colony: an acceleration approach with variable sample size. Appl Soft Comput 88:106041
Xi M, et al. (2016) Cancer feature selection and classification using a binary quantum-behaved particle swarm optimization and support vector machine. Comput Math Methods Med.
Zheng C-H et al (2008) Gene expression data classification using consensus independent component analysis. Genom Proteom Bioinform 6(2):74–82
Zhu X, Wang N (2019) Cuckoo search algorithm with onlooker bee search for modeling PEMFCs using T2FNN. Eng Appl Artif Intell 85:740–753
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Material preparation, data collection analysis and all other work were performed by Dr. Rabia Musheer Aziz.
Corresponding author
Ethics declarations
Conflict of interests
The authors have no relevant financial or non-financial interests to disclose. Also the authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Aziz, R.M. Application of nature inspired soft computing techniques for gene selection: a novel frame work for classification of cancer. Soft Comput 26, 12179–12196 (2022). https://doi.org/10.1007/s00500-022-07032-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-022-07032-9