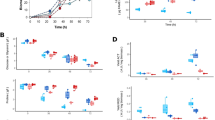
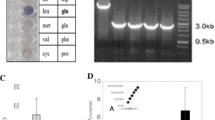
Abstract
Streptomyces, which produces many pharmaceutical antibiotics and anticancer agents, is a genus of soil-dwelling bacteria with numerous regulators that control both primary and secondary metabolism. NdgR is highly conserved in Streptomyces spp. and is known to be involved in antibiotic production, tolerance against shock and physical stress, nitrogen metabolism, leucine metabolism, and N-acetylglucosamine metabolism. As another function of NdgR, we report the involvement of NdgR in glycerol metabolism in S. coelicolor. Initially, a glycerol utilization operon containing gylCABX was found to be up-regulated in an ndgR deletion mutant (BG11) grown in N-acetylglucosamine solid minimal media compared with wild-type strain (M145). BG11 produced more antibiotics with a small amount of glycerol and increased glycerol utilization, yielding higher concentrations of lactate and acetate per cell. Moreover, fatty acid production was also changed in BG11 to produce longer chain fatty acids, phenolic compounds, alkanes, and fatty alcohols. Using a gel retardation assay, NdgR was found to bind the upstream region of gylC, working as a repressor. NdgR is a second regulator of a glycerol utilization operon, for which only one regulator, GylR was previously known.





Similar content being viewed by others
References
Hopwood DA (1999) Forty years of genetics with Streptomyces: from in vivo through in vitro to in silico. Microbiology 145(Pt 9):2183–2202
Chater KF (1993) Genetics of differentiation in Streptomyces. Annu Rev Microbiol 47:685–713
Bibb M (1996) The regulation of antibiotic production in Streptomyces coelicolor A3(2). Microbiology 142(Pt 6):1335–1344
Yang YH, Song E, Kim EJ, Lee K, Kim WS, Park SS, Hahn JS, Kim BG (2009) NdgR, an IclR-like regulator involved in amino-acid-dependent growth, quorum sensing, and antibiotic production in Streptomyces coelicolor. Appl Microbiol Biotechnol 82:501–511
Lee BR, Yi DH, Song E, Bhatia SK, Lee JH, Kim YG, Park SH, Lee YK, Kim BG, Yang YH (2015) Increased vulnerability to physical stress by inactivation of NdgR in Streptomyces coelicolor. Appl Biochem Biotechnol 175:3673–3682
Li C, Lesnik K, Liu H (2013) Microbial conversion of waste glycerol from biodiesel production into value-added products. Energies 6:4739
Clomburg JM, Gonzalez R (2013) Anaerobic fermentation of glycerol: a platform for renewable fuels and chemicals. Trends Biotechnol 31:20–28
Kurosawa K, Radek A, Plassmeier JK, Sinskey AJ (2015) Improved glycerol utilization by a triacylglycerol-producing Rhodococcus opacus strain for renewable fuels. Biotechnol Biofuels 8:31
da Silva GP, Mack M, Contiero J (2009) Glycerol: a promising and abundant carbon source for industrial microbiology. Biotechnol Adv 27:30–39
Yang F, Hanna MA, Sun R (2012) Value-added uses for crude glycerol—a byproduct of biodiesel production. Biotechnol Biofuels 5:13
Smith CP, Chater KF (1988) Structure and regulation of controlling sequences for the Streptomyces coelicolor glycerol operon. J Mol Biol 204:569–580
Hindle Z, Smith CP (1994) Substrate induction and catabolite repression of the Streptomyces coelicolor glycerol operon are mediated through the GylR protein. Mol Microbiol 12:737–745
van Wezel GP, White J, Young P, Postma PW, Bibb MJ (1997) Substrate induction and glucose repression of maltose utilization by Streptomyces coelicolor A3(2) is controlled by malR, a member of the lacl-galR family of regulatory genes. Mol Microbiol 23:537–549
Kieser T, Bibb MJ, Buttner MJ, Chater K, Hopwood DA (2000) Practical Streptomyces genetics. John Innes Centre, Norwich Research Park, Colney
Braunegg G, Sonnleitner B, Lafferty RM (1978) Rapid gas-chromatographic method for determination of poly-beta-hydroxybutyric acid in microbial biomass. Eur J Appl Microbiol Biotechnol 6:29–37
Yang YH, Brigham CJ, Budde CF, Boccazzi P, Willis LB, Hassan MA, Yusof ZA, Rha C, Sinskey AJ (2010) Optimization of growth media components for polyhydroxyalkanoate (PHA) production from organic acids by Ralstonia eutropha. Appl Microbiol Biotechnol 87:2037–2045
York GM, Lupberger J, Tian J, Lawrence AG, Stubbe J, Sinskey AJ (2003) Ralstonia eutropha H16 encodes two and possibly three intracellular poly[d-(−)-3-hydroxybutyrate] depolymerase genes. J Bacteriol 185:3788–3794
Kim SH, Lee BR, Kim JN, Kim BG (2012) NdgR, a common transcriptional activator for methionine and leucine biosynthesis in Streptomyces coelicolor. J Bacteriol 194:6837–6846
Kim JN, Jeong Y, Yoo JS, Roe JH, Cho BK, Kim BG (2015) Genome-scale analysis reveals a role for NdgR in the thiol oxidative stress response in Streptomyces coelicolor. BMC Genom 16:116
Lee B-R (2013) Transcriptional regulatory network analysis and its application for increasing antibiotics production in Streptomyces coelicolor. Seoul National University, Seoul
Nothaft H, Dresel D, Willimek A, Mahr K, Niederweis M, Titgemeyer F (2003) The phosphotransferase system of Streptomyces coelicolor is biased for N-acetylglucosamine metabolism. J Bacteriol 185:7019–7023
Nothaft H, Rigali S, Boomsma B, Swiatek M, McDowall KJ, van Wezel GP, Titgemeyer F (2010) The permease gene nagE2 is the key to N-acetylglucosamine sensing and utilization in Streptomyces coelicolor and is subject to multi-level control. Mol Microbiol 75:1133–1144
Revill WP, Bibb MJ, Hopwood DA (1996) Relationships between fatty acid and polyketide synthases from Streptomyces coelicolor A3(2): characterization of the fatty acid synthase acyl carrier protein. J Bacteriol 178:5660–5667
Revill WP, Bibb MJ, Hopwood DA (1995) Purification of a malonyltransferase from Streptomyces coelicolor A3(2) and analysis of its genetic determinant. J Bacteriol 177:3946–3952
Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36
Acknowledgements
Consulting service from the Microbial Carbohydrate Resource Bank (MCRB, Seoul, Korea) was kindly appreciated.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
The study was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2017R1D1A1B03033594 and NRF-2015M1A5A1037196), the Korea Institute of Energy Technology Evaluation and Planning (KETEP), and the Ministry of Trade, Industry and Energy (MOTIE) of the Republic of Korea (No. 20163010092150). This study was also partially supported by the Advanced Production Technology Development Program, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea (1201349190011).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lee, BR., Bhatia, S.K., Song, HS. et al. The role of NdgR in glycerol metabolism in Streptomyces coelicolor . Bioprocess Biosyst Eng 40, 1573–1580 (2017). https://doi.org/10.1007/s00449-017-1813-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-017-1813-z