Abstract
The effect of direct chemical defences in plants on the performance of insect herbivores and their natural enemies has received increasing attention over the past 10 years. However, much less is known about the scale at which this variation is generated and maintained, both within and across populations of the same plant species. This study compares growth and development of the large cabbage butterfly, Pieris brassicae, and its gregarious pupal parasitoid, Pteromalus puparum, on three wild populations [Kimmeridge (KIM), Old Harry (OH) and Winspit (WIN)] and two cultivars [Stonehead (ST), and Cyrus (CYR)] of cabbage, Brassica oleracea. The wild populations originate from the coast of Dorset, UK, but grow in close proximity with one another. Insect performance and chemical profiles were made from every plant used in the experiment. Foliar glucosinolates (GS) concentrations were highest in the wild plants in rank order WIN > OH > KIM, with lower levels found in the cultivars. Caterpillar-damaged leaves in the wild cabbages also had higher GS levels than undamaged leaves. Pupal mass in P. brassicae varied significantly among populations of B. oleracea. Moreover, development time in the host and parasitoid were correlated, even though these stages are temporally separated. Parasitoid adult dry mass closely approximated the development of its host. Multivariate statistics revealed a correlation between pupal mass and development time of P. brassicae and foliar GS chemistry, of which levels of neoglucobrassicin appeared to be the most important. Our results show that there is considerable variation in quantitative aspects of defensive chemistry in wild cabbage plants that is maintained at very small spatial scales in nature. Moreover, the performance of the herbivore and its parasitoid were both affected by differences in plant quality.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants are known to have evolved a number of direct mechanisms to defend themselves from attackers such as herbivorous insects. One of the most important of these mechanisms is the production of secondary metabolites or ‘allelochemicals’ (Rosenthal and Berenbaum 1992; Schoonhoven et al. 2005). Allelochemicals can be constitutively expressed and thus maintained at fairly constant levels in plant tissues or are induced in response to herbivore damage (Karban and Baldwin 1997; Agrawal 1999). Allelochemicals are known to act as repellents and reduce the risk of herbivores alighting and/or ovipositing on plant shoots (Schoonhoven et al. 2005). Furthermore, allelochemicals may exhibit varying degrees of toxicity and have deleterious effects on the development and survival of both specialist and generalist herbivores (Rosenthal and Berenbaum 1992; Schoonhoven et al. 2005). The development of natural enemies may also be affected by differences in plant quality, either directly through the herbivore itself or indirectly if the herbivore itself performs more poorly on certain diets (Campbell and Duffey 1981; Barbosa et al. 1991; Harvey 2005; Ode 2006).
In natural populations, it has also been shown that there is often considerable genetic variation in the expression of plant allelochemistry. This variation can be expressed at quite different local and geographical scales. For instance, several studies have reported significant qualitative and quantitative differences in levels of secondary compounds known as glucosinolates in populations of wild cabbage (Brassica oleracea) plants growing along the coast of Dorset, UK, even though they grow only a few kilometres from one another (Mithen et al. 1995; Moyes et al. 2000; Newton et al. 2009a, b). By contrast, another study found very little difference in concentrations of the main GS found in wild black mustard plants growing in the Netherlands and Sicily and which are separated by more than 1,000 km (Gols et al. 2009).
Genetic variation in allelochemistry is probably mediated by a range of biotic and abiotic selection pressures acting over different spatial and temporal scales. This variation is generated through a series of trade-offs in the expression of different traits, each of which is assumed to be costly to maintain. Consequently, in one set of conditions, one trait is favoured at the expense of other traits, whereas under a different set of conditions, another trait is favoured. In this scenario, selection pressures that vary even under small scales can generate strong local differences in the expression of certain phenotypic traits. Although differences in phenotype are frequently observed between plant populations, less attention has generally been paid to variation observed within populations. Furthermore, the effects of intra-population variation in plant quality on the behaviour and development of higher trophic levels has been little studied.
In this study, we compare development of the large cabbage white butterfly, Pieris brassicae L. (Lepidoptera: Pieridae), and its gregarious pupal parasitoid, Pteromalus puparum L. (Hymenoptera: Pteromalidae), on wild cabbage plants originating from three populations growing naturally along the coast of Dorset, UK. Like other members of the large family Brassicaceae, wild cabbage plants produce heritable secondary metabolites known as glucosinolates (GS) (Halkier and Gershenzon 2006). GS and their breakdown products have been shown to negatively affect the growth and/or development of insect herbivores (Li et al. 2000; Agrawal and Kurashige 2003; Gols et al. 2008; van Leur et al. 2008). GS are classified based on their amino acid origin into aliphatic, indole or aromatic GS (Halkier and Gershenzon 2006). In B. oleracea, the dominant GS are represented by aliphatic (derived from methionine) and indole GS (derived from tryptophan). Moreover, GS levels in plant tissues are not constant and may vary depending on the age of the plant and environmental factors (Gols et al. 2007). For instance, in wild B. oleracea, indole GS have been demonstrated to increase in response to insect herbivory, whereas foliar aliphatic GS concentrations were little affected by insect feeding damage (Gols et al. 2008).
Previous work has shown that wild cabbage plants exhibit strong inter-population variation in indole and aliphatic GS (Mithen et al. 1995; Moyes et al. 2000; Gols et al. 2008; Newton et al. 2009a, b). Cultivated cabbage is also one of the most important global crops, and the genetics and secondary chemistry of various cultivars of this plant are well described in the literature (Rosa 1999; Kushad et al. 2004). Due to the effects of artificial selection via domestication, plant breeders have often sought to reduce levels of foliar GS and to eliminate variation in various other plant traits (Gols and Harvey 2009). In spite of this, cultivated strains of cabbage have often been used in ecological studies (reviewed by Gols and Harvey 2009; Hopkins et al. 2009).
Pupal mass and larval development time of P. brassicae was measured on three wild populations and two cultivars of B. oleracea. Brood size, egg-to-adult development time and adult dry mass of Pt. puparum developing on young pupae of P. brassicae were also determined. A previous study (Harvey et al. 2007) reported that the performance of Pt. puparum and another of its hosts, the small cabbage white butterfly P. rapae, were negatively correlated with concentrations of indole GS in shoots of B. oleracea but not with total levels of GS. That study, however, examined insect performance at the population level, thus ignoring variation generated within populations of B. oleracea. In contrast, here we compare insect performance both within and between different populations of B. oleracea by obtaining development data and chemical profiles from all individual plants on which the insects were reared. Multivariate statistics are used to correlate GS chemistry with insect performance.
The main aim of the current study is to measure differences in GS concentrations in three naturally occurring B. oleracea populations from Dorset and two cultivars, and using these data to determine which GS compounds explain most of the population-related variation in GS chemistry in undamaged control plants and in plants exposed to P. brassicae feeding, respectively. The variation within and among plant populations is further analysed to explain the variation in performance of the herbivore and its parasitoid. Our two main hypotheses are that (1) there will be considerably more variation in GS profiles amongst the wild than cultivated strains of cabbage, and (2) that this will reflected in development of the herbivore and its parasitoid.
Materials and methods
Plants
Seeds of wild B. oleracea plants were collected from three populations growing adjacent to chalk cliffs along the south coast of the United Kingdom to the west and north of Swanage, Dorset. The populations are located at sites known as Old Harry (50°38′N, 1°55′E), Winspit (50°35′N, 2°02′E) and Kimmeridge (50°36′N, 2°07′E). In addition, two cultivated strains of B. oleracea were used: Brussels sprouts (var. gemmifera) cv. ‘Cyrus’, on which the laboratory culture of P. brassicae had been reared for many generations, and white cabbage (var. alba) cv. ‘Stonehead’. For simplicity we also refer to the cultivars as ‘populations’. Hereafter, the populations are abbreviated as OH (Old Harry), WIN (Winspit), KIM (Kimmeredge), CYR (Cyrus cultivar) and SH (Stonehead cultivar).
Seeds from the various cabbage populations were germinated in the last week of September 2007. Seedlings were transferred to 2.1-l pots that had been filled with potting soil (‘Lentse potgrond’ no. 4; Lent, The Netherlands). Plants were grown in facilities operated by Unifarm at Wageningen University (WU), The Netherlands, in a greenhouse compartment at 22 ± 4°C, 50–70% r.h. with a photoperiod of at least 16 h. If the light intensity dropped below a level of 500 μmol photons m2/s during the 16-h photoperiod, supplementary illumination was supplied by high pressure mercury lamps. All plants were uniformly watered on a daily basis. When plants were 4 weeks old, they were fertilized once a week with Kristallon Blauw (16N:6P:20K:3Mg) at a concentration of 2.5 mg/l. Plants were 7 weeks old at the start of the experiments. Watering and fertiliszation continued during the experiments.
Insects
Hosts and parasitoids were reared at 22 ± 2°C with a 16:8 h L:D photoperiod. Cultures of P. brassicae were obtained from insects maintained at WU, and which were originally collected from agricultural fields near the university. P. brassicae is a wide-ranging species that occurs over much of Eurasia. Larvae feed on many different brassicaceous plants, including both wild and cultivated species. Here, caterpillars were maintained on the Brussels sprouts cultivar CYR at WU. Adults of Pt. puparum were collected while ovipositing into the pupae of P. brassicae on wild cabbage plants growing in a small plot adjacent to the Institute of Ecology, Heteren, The Netherlands. Adult female wasps lay broods into the pupae of several pierid host species in nature; brood size is dependent on the size of the host (Harvey et al. 2007). Pt. puparum is an idiobiont parasitoid (i.e. it develops in non-growing hosts) and the female wasp injects permanently paralysing venom into the host preceding oviposition. In order to maximise egg production, female wasps perforate the cuticles of some hosts with their ovipositor and drink hemolymph that emerges from the wound. Proteins from host hemolymph are utilised in the production of eggs (Jervis and Kidd 1986). After hatching, the parasitoid larvae feed on internal host tissues and pupate within the cuticle of the pupa. Adult wasps emerge within a few days. To maintain parasitoid cultures in the laboratory, Pt. puparum was reared on P. brassicae pupae as described in Harvey et al. (2007). Female wasps used in the experiments were >5 days old, had been mated in rearing cages and had been given access to host pupae on one previous occasion for host-feeding purposes.
Experimental protocol: development of P. brassicae on five different B. oleracea populations
To obtain P. brassicae eggs, one plant of each of the five B. oleracea populations was placed in the rearing cage with approximately 200 butterflies in a 1:1 male-to-female sex ratio for about 2 h. After this oviposition period, each plant contained at least five egg batches. When the eggs hatched (5 days later), neonate larvae were randomly sampled from the different egg batches and transferred to new plants, seven larvae per plant, from the same population on which they originated. Plants of the different populations were randomly positioned in a greenhouse compartment with similar conditions as described above. The larvae were allowed to move and feed freely on a plant until they were 3–4 days into the final fifth instar. At this point, larvae were collected from all individual plants and transferred to plastic boxes (17 × 13 × 6 cm) with some leaf tissues from the plant on which they had been feeding previously. The boxes were checked twice a day for pupated larvae and their day of pupation was recorded. Two days later, when the cuticle had hardened, pupae were removed from the boxes and weighed on a microbalance (Sartorius CP2P; Göttingen, Germany). These pupae were subsequently offered to Pt. puparum females for parasitism (see below).
Experimental protocol: development of Pt. puparum on P. brassicae pupae that had developed on different B. oleracea populations
Pupae from the same plant were placed in pairs in Petri dishes (9.5 cm diameter) and exposed to a single female parasitoid for 24 h. After this oviposition period, the female was removed and the pupae were kept individually in dishes until adult wasp eclosion. When the wasps started to emerge, the dishes were monitored several times a day and the adult wasps were counted, sexed and transferred to labelled vials and killed by freezing. Wasps were dried in an oven at 70°C for 3 days (until constant mass) and weighed individually on a microbalance. Egg-to-adult development time was determined as the number of days between the day of oviposition and the day of adult eclosion when occurring before 1800 hours. The recorded development time for wasps that emerged after 1800 hours was extended by 1 day (the majority of the wasps emerged during the day).
Glucosinolate analysis
GS concentrations were measured in all individual plants used in the study (n = 8 from each population) in order to get a comprehensive picture of the variation that occurred both between and within populations. Leaf samples were taken from plants just after the caterpillar had been removed from them and the plants had been exposed to caterpillar feeding for approximately 20 days. To investigate the effect of P. brassicae larval feeding on foliar GS concentrations, leaf samples were also collected from undamaged control plants (n = 8) from each of the populations. Damaged and control plants were of the same age when sampled. Two discs, 17 mm diameter, were punched per leaf, one from each central part of a leaf half, and six fully unfolded leaves were sampled per plant. Leaf discs were pooled per plant in 2-ml centrifuge tubes and frozen in liquid nitrogen immediately after sampling and then stored at −80°C. Samples were later freeze-dried and pulverised. Fifty-milligram aliquots of freeze-dried material were weighed in 2-ml centrifuge tubes. GS were extracted and purified as described by van Dam et al. (2004). GS were separated on a reverse phase C-18 column (Alltima C-18, 3 μm, 150 × 4.6 mm; Alltech, Deerfield, IL, USA) on high performance liquid chromatography (HPLC; Dionex, Sunnyvale, CA, USA) with an acetonitrile–water gradient. Detection was performed with a Dionex PDA-100 photodiode array detector set to scan from 200 to 350 nm. For quantification, sinigrin (Sigma, St. Louis, MO, USA) was used as an external standard. Peaks were integrated at 229 nm for which standard response factors have been defined. The different GS compounds were identified based on their retention times and UV spectra compared to those of pure compounds sinigrin (Sigma), glucotropaeolin, glucobrassicin (kindly provided by M. Reichelt, MPI for Chemical Ecology, Jena, Germany), or compared to a certified oil seed reference (EC Community Bureau of Reference BCR-367R, Fluka, Buchs, Switzerland). GS compounds were classified based on their amino-acid origin into indole GS (derived from tryptophan), alipathic GS (derived from methionine) and aromatic GS (derived from phenylanaline) (Halkier and Gershenzon 2006).
Data analysis
To investigate the effect of plant population on the performance (pupal mass and egg-to-pupa development time) of the herbivore P. brassicae, the mean values of these parameters were calculated per plant. We used mean values to meet the assumptions of normality and equal variances which could not be achieved by transformation of measurements on individuals. Similarly, the parasitoid brood sizes per host pupa were also averaged per plant. These mean values were subjected to analysis of variance (ANOVA) with plant population as explanatory factor. Offspring sex ratios (number of males divided by total number of wasps) were also analysed by ANOVA. Here, we included mixed broods as female wasps producing only male offspring may not have been mated. Only fertilised eggs produce female offspring (Godfray 1994). Most plants had only 1–2 hosts producing mixed offspring; therefore, sex-ratio data were not averaged per plant. When the model was significant, post-hoc Tukey–Kramer multiple comparisons were conducted.
Performance data (adult mass and egg-to-adult development time) of Pt. puparum were averaged per host for male and females separately. Since the wasp data were obtained from several plants within a population, there were two sources of random error: variation among experimental units (here, mean performance per host) and variation among plants within a population. Therefore, these data were analysed with a mixed model (restricted likelihood estimation, REML). The analysis calculates Wald-type F statistics and can process different sources of variation, enabling the use of all individuals rather than using averages per plant (Piepho et al. 2003). Data were analysed with population, sex (male or female), and their interaction as fixed factors and plant as a random factor. As the number of individuals (=brood size) developing in a host may also affect the final size and or development of the emerging wasps, brood size was included as a covariate in the statistical model. Predicted means, standard errors, and individual comparisons estimated after model fitting, were used for presentation of the results. Analyses were carried out using the GenStat 12.1 software package.
Multivariate statistics (i.e. principal components analysis, PCA) were used to reveal (1) which GS compounds are important for the separation of the five cabbage populations, and (2) whether performance of the insects can be predicted based on GS present in leaf tissues on which the host has been feeding. PCA is an ordination technique that attempts to represent multivariate data in low dimensional planes, which are described by latent variables or principle components. Projection to Latent Structures Discriminant Analysis (PLS-DA) is a projection method that attempts to separate groups of observations by rotating the PCAs such that a maximum separation among classes, here B. oleracea populations, is obtained (Eriksson et al. 2006). To achieve this, a Y-data matrix of dummy variables is included which assigns a sample to its respective class. The PLS-DA extension of the SIMCA P + 12.0 software program (Umetrics, Umeå, Sweden) used for this analysis, approximates the point ‘swarm’ in X (matrix with GS variables) and Y in PLS components in such a way that maximum covariation between the components in X and Y is achieved. The results of the analysis are visualised in score plots, which reveal the sample structure according to the model components, and loading plots, which display the contribution of the variables to these components, as well as the relationships among the variables. The program’s cross validation procedure evaluates the significance of each additional component (starting with none) by comparing the goodness of fit (R 2) and the predictive value (Q 2) of the extended model with that of the reduced model. Per leaf sample, concentrations of the individual GS compounds, total GS, total aliphatic GS, total indole GS, and indole and aliphatic percentages served as variables in the model. Data were log-transformed, mean-centred and scaled to unit variance before they were subjected to the analysis. GS measured in undamaged and herbivore-damaged plant tissues were subjected to PLS-DA separately. In a second analysis, the GS chemistry data were linked to the insect performance data using PLS without discriminate analysis. The chemistry variables used in the PLS-DA were also used in this analysis. Only the GS data measured in P. brassicae-damaged plants were included. The insect performance data, which represented the Y-matrix variables, included: development time of the herbivore, pupal mass of the herbivore, biomass of male and female adult parasitoids, sex ratio of the parasitoid offspring, and parasitoid brood sizes per host.
Results
Development of P. brassicae on five different B. oleracea populations
Plant population had a significant effect on P. brassicae pupal fresh mass (F 4,35 = 10.5, P < 0.001) and egg-to-pupation development time (F 4,35 = 4.59, P = 0.004). The pupae were the heaviest on CYR plants and the lightest on KIM plants, whereas the pupae were of intermediate weight on the other three plant populations (Fig. 1a). In addition, development was prolonged on KIM plants compared to the other four plant populations (Fig. 1b).
a Pupal fresh mass and b egg-to-pupa development time of P. brassicae that were reared in a greenhouse on eight plants of either one of two cabbage cultivars (CYR or SH) or one of three wild cabbage populations (KIM, OH, WIN) from the UK (Dorset). Values are means + SE (n = 8) based on averaged values calculated per plant (initially seven caterpillars were placed on each plant); bars with the same letter are not significantly different (Tukey–Kramer tests for multiple comparisons among means with α = 0.05)
Development of Pt. puparum on P. brassicae pupae that had developed on different B. oleracea populations
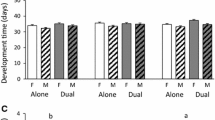
In total, 134 host pupae produced collectively 5,601 adult wasps. Brood sizes were highly variable (range 1–127; mean ± SE, 34.4 ± 4.9), but did not significantly differ in hosts that had developed on the different plant populations (F 4,35 = 0.45, P = 0.77). In addition, offspring sex ratios were similar in hosts reared on the five plant populations (F 4,94 = 0.30, P = 0.88). Population (Wald = 12.2, P = 0.03) and sex (Wald = 469, P < 0.001) also had a significant effect on adult dry mass of Pt. puparum wasps (Fig. 2a). Female wasps were significantly heavier than males. In addition, wasps were significantly smaller (based on least significant differences between means at the 5% level) when the host had developed on KIM plants compared with CYR, SH and OH plants, whereas wasps from hosts on WIN plants were of intermediate size. Brood size covaried with wasp size, with larger broods producing smaller wasps (Wald = 163, P < 0.001, slope ± SE: −4.22 ± 0.33 μg/brood size number). Population (Wald = 25.4, P < 0.001), but not sex (Wald = 2.49, P = 0.12), also influenced egg-to-adult development time (Fig. 2b). Wasps developed significantly faster on hosts from WIN plants than on hosts from the other plant populations with the exception of hosts from the CYR population on which the difference in development time was not statistically significant (based on least significant differences between means at the 5% level). Wasp development time covaried with brood size; wasps from larger broods developed faster than wasps from smaller broods (Wald = 2,411, P < 0.001: slope ± SE: −0.0787 ± 0.0051 days/brood size number).
a Adult dry mass and b egg-to-adult development time of Pt. puparum that had developed in P. brassicae pupae reared on either one of two cabbage cultivars (CYR or SH) or one of three wild cabbage populations (KIM, OH, WIN) from the UK (Dorset). Values are predicted means + SE for females (white bars) and males (grey bars) and a mean brood size of 39 based on the REML statistical model. For statistical analysis, refer to the text
When performance of P. brassicae and Pt. puparum are compared, patterns for biomass on the different plant populations were similar (Figs. 1a and 2a). In other words, larger pupae produced larger wasps for a given brood size. With respect to development, differences in development of the wasps on the various plant populations did not reflect pupal development of the herbivore (Figs. 1b and 2b). However, from the total of 242 host pupae that were initially exposed to Pt. puparum, only 134 pupae produced adult wasps. A bar graph (not shown) including only development times of the pupae that produced wasps mirrored the patterns found in Fig. 2b. Moreover, linear least square regression of wasp development times on pupal development times of the hosts revealed a significant positive correlation (F 1,131 = 889, P < 0.001, r 2 = 0.87).
Glucosinolate analyses
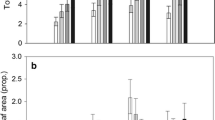
In total, 11 different GS were present in the foliage of the five B. oleracea populations: 4 indole GS (GBC, NEO, 4OH and 4MeOH), 6 aliphatic GS (IBE, SIN, GNA, PRO, epigoitrin, and glucoallyssin), and 1 aromatic GS (gluconasturtiin) (for a full description of the abbreviated names, see Fig. 3). An overview of the GS concentrations in leaf tissues of undamaged and herbivore-damaged leaf tissues (Fig. 3) shows that GS concentrations were lower in the two cultivars (CYR and SH) than in the three wild populations. In addition, in response to P. brassicae feeding the indole GS, NEO and GBC increased dramatically in the wild populations. The highest GS levels were found in WIN plants. In plants from this population, herbivory mainly resulted in quantitative changes of individual GS compounds, whereas total GS concentrations remained fairly constant. As gluconasturtiin, glucoallyssin and epirogoitrin were present in less than 5 samples and at low concentrations, they were not entered as variables in the PLS-DA analysis. The PLS-DA on GS concentrations measured in undamaged leaf tissues resulted in a model with three significant PLS components explaining 71.4% of the variation. The loading plot (Fig. 4c) shows how the GS variables relate to the patterns found in the score plot (Fig. 4a). The first component separated SH from the other four plant populations (Fig. 4a) and the variables contributing the most to this component were total indole GS (percentage and absolute value), mainly through the contribution of GBC (Fig. 4c); concentrations of GBC were much lower in leaf tissues of SH plants than in leaf tissues from plants of the other four populations. The second PLS component separated the other four populations. The populations aligned along the second component in the following order: CYR, KIM, OH and WIN. IBE and total GS concentrations, mainly through the aliphatic GS GNA, were the most important variables contributing to the second component`. CYR plants contained relatively high levels of IBE, whereas WIN plants contained relatively high overall levels of aliphatic GS dominated by GNA. Concentrations of IBE and GNA were intermediate in OH and KIM plants.
GS concentrations in leaf tissues of undamaged control (left bars in each graph) and P. brassicae-damaged (right bars in each graph) plant individuals sampled from two cabbage cultivars, a CYR and b SH and three wild cabbage populations originating from the UK (Dorset), c KIM, d WIN and e OH, respectively. Plants are ordered from low to high total GS concentrations. Samples from control and damaged plant tissues were taken from different plant individuals. The order of the compounds in the bars is the same as the order of the compounds in the legend. The GS compounds (abbreviation and scientific name) were classified as indole GS: glucobrassicin (GBC 3-indolylmethyl GS), neoglucobrassicin (NEO 1-methoxy-3-indolylmethyl GS), 4-hyddroxy glucobrassicin (4OH 4-hydroxy-3-indolylmethyl GS), 4-methoxy glucobrassicin (4MeOH 4-methoxy-3-indolylmethyl GS) and as aliphatic GS: glucoiberin (IBE 3-methylsulfinylpropyl GS), sinigrin (SIN 2-propenyl GS), glucoraphinin (RAPH 4-methylsulfiniyl-3-butenyl GS), gluconapin (GNA 3-butenyl GS), progoitrin [PRO 2(R)-2-hydroxy-3-butenyl GS]
PLS-DA (Projection to Latent Structures Discriminant Analysis) of 14 GS related variables measured in undamaged (a, c) and P. brassicae-damaged (b, d) leaf tissues sampled from two cabbage cultivars (CYR and SH , open symbols) and three wild cabbage populations (KIM, OH, WIN, closed symbols) originating from the UK (Dorset). The score plots (a, b) visualise the structure of the samples according to the first two PLS components with the explained variance in parentheses. The ellipse in the score plots defines the Hotelling’s T2 confidence region, which is a multivariate generalisation of Student’s t test and provides a 95% confidence interval for the observations. The loading plots (c, d) define the orientation of the PLS planes with the original variables in the X (GS variables) and Y space (‘class’ variables), respectively. They reveal the magnitude and direction of correlation of the original variables with the first two PLS components. For an explanation of the GS abbreviations, see Fig. 3. Total GS, total indole GS (ind gs) total aliphatic GS (ali gs) and their percentages of the total amount (%ind and %ali, respectively) were also included as variables. Model statistics for undamaged leaves with three significant components: R 2 X = 0.73, R 2 Y = 0.64, Q 2 = 0.61, and damaged leaves with four significant componenets, R 2 X = 0.85, R 2 Y = 0.82, Q 2 = 0.76)
A PLS-DA on GS concentrations found in P. brassicae-damaged leaf tissues resulted in a model with four significant PLS components explaining 85% of the variation and revealed a different separation pattern compared to undamaged control plants (Fig. 4a, c). The first PLS component aligned the populations in the following order KIM < OH = WIN < CYR < SH and the second component further separated CYR from SH and KIM from the other two wild populations OH and WIN. The wild populations (KIM, OH and WIN) contained relatively high levels of indole GS (Fig. 4d) compared to the two cultivars SH and CYR. In OH and WIN, the dominating indole GS was GBC, whereas in KIM this was NEO (Fig. 4d).
Fitting a PLS model through the two datasets, GS and performance data, respectively, produced a model with three significant components explaining 60% of the variation among the GS variables (= R 2 X), 16% among the performance variables (= R 2 Y), and 11% of the predicted variation (= Q 2). Pupal mass and development time of the host, which were negatively correlated, were the only two performance variables that significantly contributed to the PLS model based on the programs’ cross-validation procedure. A reduced model with only pupal mass and development time in the Y-matrix increased the goodness of fit (R 2 Y = 0.34) and the predictive ability (Q 2 = 0.31) of the PLS model. The most important GS variables for explaining variation in the performance parameters were 4OH, NEO, and % aliphatic GS. Development time of the herbivore correlated positively with levels of 4OH and NEO, and negatively with % aliphatic GS, whereas the opposite pattern was found for pupal mass. Pupae were smaller and extended their development on KIM plants which have the highest levels of NEO and lowest percentages of aliphatic GS in their leaf tissues. Concentrations of 4OH tended to be low (<0.2 μmol/g DW) and there was no clear relationship between levels of this compound and plant population.
Discussion
The results of this investigation reveal that the development of the large cabbage white butterfly P. brassicae and its gregarious pupal parasitoid Pt. puparum differed across different cultivated and wild populations of B. oleracea. Although brood size and sex ratio were fairly constant in Pt. puparum, both host pupal mass and parasitoid adult mass were smallest when developing on KIM plants. Development time in P. brassicae was also extended when caterpillars were reared on plants from the KIM population. Development time of the parasitoid was correlated positively with development time of the host; this is interesting because the host does not grow during parasitism and thus some aspect of host quality is passed from the host to the parasitoid in such as way as to affect its rate of development. Overall, these results show that the development of the parasitoid and its host are well integrated across a spectrum of fitness-related traits (Harvey 2005).
There were significant differences in chemical profiles of wild and domesticated cabbage plants that were exhibited both within and between populations, although the latter was more pronounced. Furthermore, analyses of GS chemistry revealed that aliphatic GS dominate the profile in undamaged leaf tissues of B. oleracea, whereas indole GS become more prevalent in herbivore-damaged leaf tissues. Thus, aliphatic GS tend to be constitutively expressed, whereas indole GS tend to be induced in response to herbivory. We also found very little intra-population (or strain) variation in the cultivars CYR and SH but significantly more in the wild populations. Furthermore, the amount of induction and total levels of GS were considerably higher in the wild cabbage populations, presumably due to the effects of artificial selection in domestic strains that have been aimed at emphasising certain traits (e.g. vegetative growth) while reducing others (e.g. the production of secondary plant compounds) (Evans 1993; Rosenthal and Dirzo 1997). Of particular interest is the fact that GS profiles in the wild cabbages differed profoundly even when the plants from different populations grew in close proximity. Multivariate analysis revealed that KIM plants were quite dissimilar from the other two wild populations, even though KIM plants grow only about 7 km from the WIN population; at the same time, the WIN and OH populations, which exhibited more similarity in their GS profiles, are separated by almost 15 km. This shows that local conditions in which the plants grow can generate highly divergent chemotypes. Individual cabbage plants can live over 20 years in the wild and populations of this species growing along the Dorset coastline appear to have been stable for decades and perhaps centuries (Wichmann et al. 2008), reinforcing the potential importance of local adaptation to a suite of local selection pressures.
Using multivariate statistics we also found that there was a link between GS chemistry in B. oleracea and performance parameters of the herbivore P. brassicae. These effects appeared to become more diffuse up the food chain. Pupal mass was negatively correlated with concentrations of the indole GS, neoglucobrassicin (NEO), which was highest in plants from the KIM population. Recent work (Gols et al. 2008) has found that the performance of P. rapae and its koinobiont endoparasitoid, Cotesia rubecula, are also negatively affected when developing on KIM plants that exhibit high NEO expression. Previous studies have reported that indole GS also negatively affect the development of the green peach aphid Myzus persicae (Kim and Jander 2007). By contrast, high levels of aliphatic GS and not indole GS were negatively correlated with the performance of the generalist herbivore Mamestra brassicae (Gols et al. 2008). This reveals that different types of GS have evolved to hamper the performance of specialist and generalist herbivores, potentially owing to the fact that these herbivores possess different strategies in dealing with plant allelochemicals. For instance, P. rapae larvae possess ‘nitrile specifier proteins’ (NSP) in their gut that convert toxic isothiocyantes into harmless nitriles which are then excreted in their frass (Wittstock et al. 2004). Detoxification strategies of generalist herbivores are often much less evolutionarily refined than those found in specialist herbivores (Krieger et al. 1971; Johnson 1999).
In recent years, there has been a renewed interest in the effects of plant quality on the development of higher trophic levels, with much focus being placed on parasitoids and hyperparasitoids in the third and fourth trophic levels (Harvey et al. 2003; Bukovinszky et al. 2008; Harri et al. 2008; Morse 2009). The effects of plant allelochemistry on natural enemy fitness are often strongly correlated with the effects on the herbivore host itself. That is, when the host develops poorly on a specific diet, higher trophic levels associated with that host also perform poorly (Barbosa et al. 1986; Gunasena et al. 1990; Harvey 2005).
One important factor determining the susceptibility of parasitoids to variations in plant quality is in the way that the parasitoid exploits host resources. Koinobiont parasitoids attack hosts that continue feeding and growing during the parasitism phase (Askew and Shaw 1986). Adult female koinobionts may be much larger than the host when it is parasitised meaning that their fitness and that of their progeny is dependent on how fast and large the host grows during parasitism. Koinobiont parasitoids are thus unable to predict future host quality and are dependent on how well the host responds to its food plant. Alternatively, idiobiont parasitoids attack non-feeding/growing host stages, such as pupae (Askew and Shaw 1986), and thus their development is dependent on the host’s nutritional history prior to parasitism. In gregarious idiobionts, it is known that female wasps adjust their clutch size in response to differences in host size as a means of stabilising the amount of resources available to their progeny (Godfray 1994). Larger broods are laid on larger hosts and vice versa. The results of this study suggest that Pt. puparum is affected by differences in the quality of host resources as well as by their total amount. This kind of effect has rarely been reported in idiobiont parasitoids, and reveals that something in the host diet is transferred from the caterpillars to their pupae, and then on to the parasitoids developing on these pupae. Given that P. brassicae does not sequester GS during its development (Müller et al. 2003), it would be interesting to determine what is stored in its tissues that affects the development of Pt. puparum.
In summary, this study has reported that there is significant between- as well as within-population variation in the types and levels of GS present in shoots of wild B. oleracea plants, as well as in the levels of induction after herbivore damage. Furthermore, the development of a specialist herbivore, P. brassicae, and its pupal parasitoid, Pt. puparum, were correlated not so much with total GS concentrations as with the amount of the indole GS NEO in their diet. Thus, although WIN plants produced the highest total GS concentrations, plants from the KIM population, in producing the highest levels of NEO, were of lower quality for herbivore and parasitoid performance. GS profiles of the cultivated plants were very different from those of the wild types, probably due to the effects of domestication, and we argue that this makes them poor models for studies of direct chemical defenses in plants. The fact that there were considerable within-population differences in the expression of GS in the wild cabbages reinforces the view of Newton et al. (2009b) that there are likely divergent selection pressures maintaining this variation. Further studies are aimed at exploring the population genetics of herbivores and their parasitoids associated with the wild cabbages growing along the coasts of England and France in order to determine if they exhibit local adaptation to the different chemical profiles found in meta-populations of B. oleracea.
References
Agrawal AA (1999) Induced plant defense: evolution of induction and adaptive phenotypic plasticity. In: Agrawal AA, Tuzun S, Bent E (eds) Induced plant defenses against pathogens and herbivores: biochemistry, ecology and agriculture. APS, Minosota, pp 251–268
Agrawal AA, Kurashige NS (2003) A role for isothiocyanates in plant resistance against the specialist herbivore Pieris rapae. J Chem Ecol 29:1403–1415
Askew RR, Shaw MR (1986) Parasitoid communities: their size, structure, and development. In: Waage J, Greathead D (eds) Insect parasitoids. Academic, London, pp 225–264
Barbosa P, Saunders JA, Kemper J, Trumbule R, Olechno J, Martinat P (1986) Plant allelochemicals and insect parasitoids: effects of nicotine on Cotesia congregata (Say) (Hymenoptera, Braconidae) and Hyposoter annulipes (Cresson) (Hymenoptera, Ichneumonidae). J Chem Ecol 12:1319–1328
Barbosa P, Gross P, Kemper J (1991) Influence of plant allelochemicals on the tobacco hornworm and its parasitoid, Cotesia congregata. Ecology 72:1567–1575
Bukovinszky T, van Veen FJF, Jongema Y, Dicke M (2008) Direct and indirect effects of resource quality on food web structure. Science 319:804–807
Campbell BC, Duffey SS (1981) Alleviation of α-tomatine-induced toxicity to the parasitoid, Hyposoter exiguae, by phytosterols in the diet of the host, Heliothis zea. J Chem Ecol 7:927–946
Evans LT (1993) Crop evolution, adaptation and yield. Cambridge University Press, New York
Godfray HCJ (1994) Parasitoids. Behavioral and evolutionary ecology. Princeton University Press, New Jersey
Gols R, Harvey JA (2009) Plant-mediated effects in the Brassicaceae on the performance and behaviour of parasitoids. Phytochem Rev 8:187–206
Gols R, Raaijmakers CE, van Dam NM, Dicke M, Bukovinszky T, Harvey JA (2007) Temporal changes affect plant chemistry and tritrophic interactions. Basic Appl Ecol 8:421–433
Gols R, Wagenaar R, Bukovinszky T, van Dam NM, Dicke M, Bullock JM, Harvey JA (2008) Genetic variation in defense chemistry in wild cabbages affects herbivores and their endoparasitoids. Ecology 89:1616–1626
Gols R, van Dam NM, Raaijmakers CE, Dicke M, Harvey JA (2009) Are population differences in plant quality reflected in the preference and performance of two endoparasitoid wasps? Oikos 118:733–743
Gunasena GH, Vinson SB, Williams HJ (1990) Effects of nicotine on growth, development, and survival of the tobacco budworm (Lepidoptera, Noctuidae) and the parasitoid Campoletis sonorensis (Hymenoptera, Ichneumonidae). J Econ Entomol 83:1777–1782
Halkier BA, Gershenzon J (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57:303–333
Harri SA, Krauss J, Muller CB (2008) Fungal endosymbionts of plants reduce lifespan of an aphid secondary parasitoid and influence host selection. Proc R Soc Lond B 275:2627–2632
Harvey JA (2005) Factors affecting the evolution of development strategies in parasitoid wasps: the importance of functional constraints and incorporating complexity. Entomol Exp Appl 117:1–13
Harvey JA, van Dam NM, Gols R (2003) Interactions over four trophic levels: foodplant quality affects development of a hyperparasitoid as mediated through a herbivore and its primary parasitoid. J Anim Ecol 72:520–531
Harvey JA, Gols R, Wagenaar R, Bezemer TM (2007) Development of an insect herbivore and its pupal parasitoid reflect differences in direct plant defense. J Chem Ecol 33:1556–1569
Hopkins RJ, van Dam NM, van Loon JJA (2009) Role of glucosinolates in insect–plant relationships and multitrophic interactions. Annu Rev Entomol 54:57–83
Jervis MA, Kidd NAC (1986) Host-feeding strategies in hymenopteran parasitoids. Biol Rev 61:395–434
Johnson KS (1999) Comparative detoxification of plant (Magnolia virginiana) allelochemicals by generalist and specialist Saturniid silkmoths. J Chem Ecol 25:253–269
Karban R, Baldwin IT (1997) Induced responses to herbivory. University of Chicago Press, Chicago
Kim JH, Jander G (2007) Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant J 49:1008–1019
Krieger RI, Feeny PP, Wilkinson CF (1971) Detoxication enzymes in guts of caterpillars: an evolutionary answer to plant defenses. Science 172:579–581
Kushad MM, Cloyd R, Babadoost MB (2004) Distribution of glucosinolates in ornamental cabbage and kale cultivars. Sci Hortic 101:215–221
Li Q, Eigenbrode SD, Stringham GR, Thiagarajah MR (2000) Feeding and growth of Plutella xylostella and Spodoptera eridania on Brassica juncea with varying glucosinolate concentrations and myrosinase activities. J Chem Ecol 26:2401–2419
Mithen R, Raybould AF, Giamoustaris A (1995) Divergent selection for secondary metabolites between wild populations of Brassica oleracea and its implications for plant-herbivore interactions. Heredity 75:472–484
Morse DH (2009) Four-level interactions: herbivore use of ferns and subsequent parasitoid-hyperparasitoid performance. Ecol Entomol 34:246–253
Moyes CL, Collin HA, Britton G, Raybould AE (2000) Glucosinolates and differential herbivory in wild populations of Brassica oleracea. J Chem Ecol 26:2625–2641
Müller C, Agerbirk N, Olsen CE (2003 Lack of sequestration of host plant glucosinolates in Pieris rapae and P. grarricae. Chemoecology 13:47–54
Newton E, Bullock JM, Hodgson D (2009a) Bottom-up effects of glucosinolate variation on aphid colony dynamics in wild cabbage populations. Ecol Entomol 34:614–623
Newton EL, Bullock JM, Hodgson DJ (2009b) Glucosinolate polymorphism in wild cabbage (Brassica oleracea) influences the structure of herbivore communities. Oecologia 160:63–76
Ode PJ (2006) Plant chemistry and natural enemy fitness: effects on herbivore and natural enemy interactions. Annu Rev Entomol 51:163–185
Piepho HP, Buchse A, Emrich K (2003) A hitchhiker’s guide to mixed models for randomized experiments. J Agron Crop Sci 189:310–322
Rosa EAS (1999) Chemical composition. In: Gomez-Campo C (ed) Developments in plant genetics and breeding, 4. Biology of Brassica coenospecies. Elsevier, Amsterdam, pp 315–357
Rosenthal GA, Berenbaum MR (1992) Herbivores: their interactions with secondary plant metabolites. Academic, San Diego
Rosenthal JP, Dirzo R (1997) Effects of life history, domestication and agronomic selection on plant defence against insects: evidence from maizes and wild relatives. Evol Ecol 11:337–355
Schoonhoven LM, van Loon JJA, Dicke M (2005) Insect–plant biology, 2nd edn. Oxford University Press, UK
Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, CW, Wold S (2006) Multi- and megavariate data analysis. Part 1: basic principles and applications. Umetrics AB, Umeå
van Dam NM, Witjes L, Svatoš A (2004) Interactions between aboveground and belowground induction of glucosinolates in two wild Brassica species. New Phytol 161:801–810
van Leur H, Vet L, van der Putten W, van Dam N (2008) Barbarea vulgaris glucosinolate phenotypes differentially affect performance and preference of two different species of lepidopteran herbivores. J Chem Ecol 34:121–131
Wichmann MC, Alexander MJ, Hails RS, Bullock JM (2008) Historical distribution and regional dynamics of two Brassica species. Ecography 31:673–684
Wittstock U, Agerbirk N, Stauber EJ, Olsen CE, Hippler M, Mitchell-Olds T, Gershenson J, Vogel H (2004) Successful herbivore attack due to metabolic diversion of a plant chemical defense. Proc Natl Acad Sci USA 101:4859–4864
Acknowledgments
The authors wish to thank André Gidding and Leo Koopman for rearing P. brassicae and Roel Wagenaar for rearing Pt. puparum.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Diethart Matthies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Harvey, J.A., van Dam, N.M., Raaijmakers, C.E. et al. Tri-trophic effects of inter- and intra-population variation in defence chemistry of wild cabbage (Brassica oleracea). Oecologia 166, 421–431 (2011). https://doi.org/10.1007/s00442-010-1861-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00442-010-1861-4