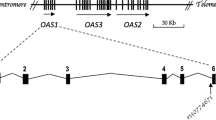
Abstract
Mutations in the GJB2 gene are known to be a major cause of autosomal recessive deafness 1A (OMIM 220290). The most common pathogenic variants of the GJB2 gene have a high ethno-geographic specificity in their distribution, being attributed to a founder effect related to the Neolithic migration routes of Homo sapiens. The c.-23 + 1G > A splice site variant is frequently found among deaf patients of both Caucasian and Asian origins. It is currently unknown whether the spread of this mutation across Eurasia is a result of the founder effect or if it could have multiple local centers of origin. To determine the origin of c.-23 + 1G > A, we reconstructed haplotypes by genotyping SNPs on an Illumina OmniExpress 730 K platform of 23 deaf individuals homozygous for this variant from different populations of Eurasia. The analyses revealed the presence of common regions of homozygosity in different individual genomes in the sample. These data support the hypothesis of the common founder effect in the distribution of the c.-23 + 1G > A variant of the GJB2 gene. Based on the published data on the c.-23 + 1G > A prevalence among 16,177 deaf people and the calculation of the TMRCA of the modified f2-haplotypes carrying this variant, we reconstructed the potential migration routes of the carriers of this mutation around the world. This analysis indicates that the c.-23 + 1G > A variant in the GJB2 gene may have originated approximately 6000 years ago in the territory of the Caucasus or the Middle East then spread throughout Europe, South and Central Asia and other regions of the world.





Similar content being viewed by others
Code availability
Bioinformatic analyses were done with publicly available software packages as described in the methods. Custom codes to aid in variant filtering are available upon request.
References
Abidi O, Boulouiz R, Nahili H, Imken L, Rouba H, Chafik A, Barakat A (2008) The analysis of three markers flanking GJB2 gene suggests a single origin of the most common 35delG mutation in the Moroccan population. Biochem Biophys Res Commun 377(3):971–974. https://doi.org/10.1016/j.bbrc.2008.10.086
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19(9):1655–1664. https://doi.org/10.1101/gr.094052.109
Balci B, Gerçeker FO, Aksoy S, Sennaroğlu G, Kalay E, Sennaroğlu L, Dinçer P (2005) Identification of an ancestral haplotype of the 35delG mutation in the GJB2 (connexin 26) gene responsible for autosomal recessive non-syndromic hearing loss in families from the Eastern Black Sea Region in Turkey. Turk J Pediatr 47(3):213–221
Barashkov NA, Dzhemileva LU, Fedorova SA, Teryutin FM, Posukh OL, Fedotova EE, Lobov SL, Khusnutdinova EK (2011) Autosomal recessive deafness 1A (DFNB1A) in Yakut population isolate in Eastern Siberia: extensive accumulation of the splice site mutation IVS1+1G>A in GJB2 gene as a result of founder effect. J Hum Genet 56(9):631–639. https://doi.org/10.1038/jhg.2011.72
Behar DM, Yunusbayev B, Metspalu M, Metspalu E, Rosset S, Parik J, Rootsi S, Chaubey G, Kutuev I, Yudkovsky G, Khusnutdinova EK, Balanovsky O, Semino O, Pereira L, Comas D, Gurwitz D, Bonne-Tamir B, Parfitt T, Hammer MF, Skorecki K, Villems R (2010) The genome-wide structure of the Jewish people. Nature 466(7303):238–242. https://doi.org/10.1038/nature09103
Behar DM, Metspalu M, Baran Y, Kopelman NM, Yunusbayev B, Gladstein A, Tzur S, Sahakyan H, Bahmanimehr A, Yepiskoposyan L, Tambets K, Khusnutdinova E, Kushniarevich A, Balanovsky O, Balanovsky E, Kovacevic L, Marjanovic D, Mihailov E, Kouvatsi A, Triantaphyllidis C, King RJ, Semino O, Antonio Torroni A, Hammer MF, Metspalu E, Skorecki K, Rosset S, Halperin E, Villems R, Rosenberg NA (2013) No evidence from genome-wide data of a Khazar origin for the Ashkenazi Jews. Hum Biol 85(6):859–900. https://doi.org/10.3378/027.085.0604
Bliznetz EA, Lalayants MR, Markova TG, Balanovsky OP, Balanovska EV, Skhalyakho RA, Pocheshkhova EA, Nikitina NV, Voronin SV, Kudryashova EK, Glotov OS, Polyakov AV (2017) Update of the GJB2/DFNB1 mutation spectrum in Russia: a founder Ingush mutation del(GJB2-D13S175) is the most frequent among other large deletions. J Hum Genet 62(8):789–795. https://doi.org/10.1038/jhg.2017.42
Carranza C, Menendez I, Herrera M, Castellanos P, Amado C, Maldonado F, Rosales L, Escobar N, Guerra M, Alvarez D, Foster J 2nd, Guo S, Blanton SH, Bademci G, Tekin M (2015) A Mayan founder mutation is a common cause of deafness in Guatemala. Clin Genet 89(4):461–465. https://doi.org/10.1111/cge.12676
Chan DK, Chang KW (2014) GJB2-associated hearing loss: systematic review of worldwide prevalence, genotype, and auditory phenotype. Laryngoscope 124(2):E34-53. https://doi.org/10.1002/lary.24332
del Castillo FJ, del Castillo I (2017) DFNB1 non-syndromic hearing impairment: diversity of mutations and associated phenotypes. Front Mol Neurosci 10:428. https://doi.org/10.3389/fnmol.2017.00428
Denoyelle F, Marlyn S, Weil D, Moatti D, Chauvin P, Garabédian EN, Petit C (1999) Clinical features of the prevalent form of childhood deafness, DFNB1, due to a Connexin–26 gene defect: implications for genetic counseling. Lancet 353(9161):1298–1303. https://doi.org/10.1016/S0140-6736(98)11071-1
Dror AA, Avraham KB (2009) Hearing loss: mechanisms revealed by genetics and cell biology. Annu Rev Genet 43:411–437. https://doi.org/10.1146/annurev-genet-102108-134135
Dzhemileva LU, Posukh OL, Barashkov NA, Fedorova SA, Teryutin FM, Akhmetova VL, Khidiyatova IM, Khusainova RI, Lobov SL, Khusnutdinova EK (2011) Haplotype diversity and reconstruction of ancestral haplotype associated with the c.35delG mutation in the GJB2 (Cx26) gene among the volgo-ural populations of Russia. Acta Naturae 3(3):52–63
Erdenechuluun J, Lin YH, Ganbat K, Bataakhuu D, Makhbal Z, Tsai CY, Lin YH, Chan YH, Hsu CJ, Hsu WC, Chen PL, Wu CC (2018) Unique spectra of deafness–associated mutations in Mongolians provide insights into the genetic relationships among Eurasian populations. PLoS ONE 13(12):e0209797. https://doi.org/10.1371/journal.pone.0209797
Fedorova SA, Reidla M, Metspalu E, Metspalu M, Rootsi S, Tambets K, Trofimova N, Zhadanov SI, Kashani BH, Olivieri A, Voevoda MI, Osipova LP, Platonov FA, Tomsky MI, Khusnutdinova EK, Torroni A, Villems R (2013) Autosomal and uniparental portraits of the native populations of Sakha (Yakutia): implications for the peopling of Northeast Eurasia. BMC Evol Biol 13:127. https://doi.org/10.1186/1471-2148-13-127
Gallant E, Francey L, Tsai EA, Berman M, Zhao Y, Fetting H, Kaur M, Deardorff MA, Wilkens A, Clark D, Hakonarson H, Rehm HL, Krantz ID (2013) Homozygosity for the V37I GJB2 mutation in fifteen probands with mild to moderate sensorineural hearing impairment: further confirmation of pathogenicity and haplotype analysis in Asian populations. Am J Med Genet A 161A(9):2148–2157. https://doi.org/10.1002/ajmg.a.36042
Green GE (1999) Carrier rates in the midwestern United States for GJB2 mutations causing inherited deafness. JAMA 281(23):2211. https://doi.org/10.1001/jama.281.23.2211
Kokotas H, Van Laer L, Grigoriadou M, Iliadou V, Economides J, Pomoni S, Pampanos A, Eleftheriades N, Ferekidou E, Korres S, Giannoulia-Karantana A, Van Camp G, Petersen MB (2008) Strong linkage disequilibrium for the frequent GJB2 35delG mutation in the Greek population. Am J Med Genet A 146A(22):2879–2884. https://doi.org/10.1002/ajmg.a.32546
Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM (2008) Worldwide human relationships inferred from genome-wide patterns of variation. Science 319(5866):1100–1104. https://doi.org/10.1126/science.1153717
Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM (2010) Robust relationship inference in genome-wide association studies. Bioinformatics 26(22):2867–2873. https://doi.org/10.1093/bioinformatics/btq559
Mathieson I, McVean G (2014) Demography and the age of rare variants. PLoS Genet 10(8):e1004528. https://doi.org/10.1371/journal.pgen.1004528
Metspalu M, Romero IG, Yunusbayev B, Chaubey G, Mallick CB, Hudjashov G, Nelis M, Mägi R, Metspalu E, Remm M, Pitchappan R, Singh L, Thangaraj K, Villems R, Kivisild T (2011) Shared and unique components of human population structure and genome-wide signals of positive selection in South Asia. Am J Med Genet 89(6):731–744. https://doi.org/10.1016/j.ajhg.2011.11.010
Morell RJ, Kim HJ, Hood LJ, Goforth L, Friderici K, Fisher R, Van Camp G, Berlin CI, Oddoux C, Ostrer H, Keats B, Friedman TB (1998) Mutations in the connexin 26 gene (GJB2) among Ashkenazi Jews with nonsyndromic recessive deafness. N Engl J Med 339(21):1500–1505. https://doi.org/10.1056/NEJM199811193392103
Norouzi V, Azizi H, Fattahi Z, Esteghamat F, Bazazzadegan N, Nishimura C, Nikzat N, Jalalvand K, Kahrizi K, Smith RJH, Najmabadi H (2011) Did the GJB2 35delG mutation originate in Iran? Am J Med Genet A 155A(10):2453–2458. https://doi.org/10.1002/ajmg.a.34225
Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. Plos Genet 2(12):e190. https://doi.org/10.1371/journal.pgen.0020190
Posukh OL, Zytsar MV, Bady-Khoo MS, Danilchenko VY, Maslova EA, Barashkov NA, Bondar AA, Morozov IV, Maximov VN, Voevoda MI (2019) Unique mutational spectrum of the GJB2 gene and its pathogenic contribution to deafness in Tuvinians (Southern Siberia, Russia): a high prevalence of rare variant c.516G>C (p.Trp172Cys). Genes (basel) 10(6):429. https://doi.org/10.3390/genes10060429
Raghavan M, Skoglund P, Graf K, Metspalu M, Albrechtsen A, Moltke I, Rasmussen S, Stafford TW Jr, Orlando L, Metspalu E, Karmin M, Tambets K, Rootsi S, Mägi R, Campos PF, Balanovska E, Balanovsky O, Khusnutdinova E, Litvinov S, Osipova LP, Fedorova SA, Voevoda MI, DeGiorgio M, Sicheritz-Ponten T, Brunak S, Demeshchenko S, Kivisild T, Villems R, Nielsen R, Jakobsson M, Willerslev E (2013) No evidence from genome-wide data of a Khazar Origin for the Ashkenazi Jews. Hum Biol 85(6):859–900. https://doi.org/10.3378/027.085.0604
RamShankar M, Girirajan S, Dagan O, Ravi Shankar HM, Jalvi R, Rangasayee R, Avraham KB, Anand A (2003) Contribution of connexin26 (GJB2) mutations and founder effect to non-syndromic hearing loss in India. J Med Genet 40(5):e68. https://doi.org/10.1136/jmg.40.5.e68
Rasmussen M, Li Y, Lindgreen S, Pedersen JS, Albrechtsen A, Moltke I, Metspalu M, Metspalu E, Kivisild T, Gupta R, Bertalan M, Nielsen K, Gilbert MTP, Wang Y, Raghavan M, Campos PF, Kamp HM, Wilson Andrew S, Gledhill Andrew R, Tridico S, Bunce M, Lorenzen ED, Binladen J, Guo X, Zhao J, Zhang X, Zhang H, Li Z, Chen M, Orlando L, Kristiansen K, Bak M, Tommerup N, Bendixen C, Pierre TL, Gronnow B, Meldgaard M, Andreasen C, Fedorova SA, Osipova LP, Higham TFG, Ramsey CB, TvO H, Nielsen FC, Crawford MH, Brunak S, Sicheritz-Ponten T, Villems R, Nielsen R, Krogh A, Wang J, Willerslev E (2010) Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature 463(7282):757–762. https://doi.org/10.1038/nature08835
Rothrock CR, Murgia A, Sartorato EL, Leonardi E, Wei S, Lebeis SL, Yu LE, Elfenbein JL, Fisher RA, Friderici KH (2003) Connexin 26 35delG does not represent a mutational hotspot. Hum Genet 113(1):18–23. https://doi.org/10.1007/s00439-003-0944-2
Shahin H, Walsh T, Sobe T, Lynch E, King MC, Avraham KB, Kanaan M (2002) Genetics of congenital deafness in the Palestinian population: multiple connexin 26 alleles with shared origins in the Middle East. Hum Genet 110(3):284–289. https://doi.org/10.1007/s00439-001-0674-2
Shinagawa J, Moteki H, Nishio SY, Noguchi Y, Usami SI (2020) Haplotype analysis of GJB2 mutations: Founder effect or mutational hot spot? Genes (basel) 11(3):250. https://doi.org/10.3390/genes11030250
Solovyev AV, Barashkov NA, Bady-Khoo MS, Zytsar MV, Posukh OL, Romanov GP, Rafailov AM, Sazonov NN, Alexeev AN, Dzhemileva LU, Khusnutdinova EK, Fedorova SA (2017) Reconstruction of SNP haplotypes with mutation c.-23+1G>A in human gene GJB2 (Chromosome 13) in some populations of Eurasia. Russ J Genet 53:936. https://doi.org/10.1134/S1022795417080099
Stenson PD, Mort M, Ball EV, Chapman M, Evans K, Azevedo L, Hayden M, Heywood S, Millar DS, Phillips AD, Cooper DN (2020) The Human Gene Mutation Database (HGMD®): optimizing its use in a clinical diagnostic or research setting. Hum Genet 139(10):1197–1207. https://doi.org/10.1007/s00439-020-02199-3
Tassi F, Vai S, Ghirotto S, Lari M, Modi A, Pilli E, Brunelli A, Susca RR, Budnik A, Labuda D, Alberti F, Lalueza-Fox C, Reich D, Caramelli D, Barbujani G (2017) Genome diversity in the Neolithic Globular Amphorae culture and the spread of Indo-European languages. Proc R Soc 284(1867):20171540. https://doi.org/10.1098/rspb.2017.1540
Tekin M, Xia XJ, Erdenetungalag R, Cengiz FB, White TW, Radnaabazar J, Dangaasuren B, Tastan H, Nance WE, Pandya A (2010) GJB2 mutations in Mongolia: complex alleles, low frequency, and reduced fitness of the deaf. Ann Hum Genet 74(2):155–164. https://doi.org/10.1111/j.1469-1809.2010.564.x
The 1000 Genomes Project Consortium (2015) A global reference for human genetic variation. Nature 526(7571):68–74. https://doi.org/10.1038/nature15393
Tsukada K, Nishio S, Hattori M, Usami S (2015) Ethnic-specific spectrum of GJB2 and SLC26A4 mutations: their origin and a literature review. Ann Otol Rhinol Laryngol 124(1 suppl):61S-76S. https://doi.org/10.1177/0003489415575060
Van Laer L, Coucke P, Mueller RF, Caethoven G, Flothmann K, Prasad SD, Chamberlin GP, Houseman M, Taylor GR, Van de Heyning CM, Fransen E, Rowland J, Cucci RA, Smith RJ, Van Camp G (2001) A common founder for the 35delG GJB2 gene mutation in connexin 26 hearing impairment. J Med Genet 38(8):515–518. https://doi.org/10.1136/jmg.38.8.515
Yan D, Park HJ, Ouyang XM, Pandya A, Doi K, Erdenetungalag R, Du LL, Matsushiro N, Nance WE, Griffith AJ, Liu XZ (2003) Evidence of a founder effect for the 235delC mutation of GJB2 (connexin 26) in East Asians. Hum Genet 114(1):44–50. https://doi.org/10.1007/s00439-003-1018-1
Yunusbayev B, Metspalu M, Järve M, Kutuev I, Rootsi S, Metspalu E, Behar DM, Varendi K, Sahakyan H, Khusainova R, Yepiskoposyan L, Khusnutdinova EK, Underhill PA, Kivisild T, Villems R (2012) The Caucasus as an asymmetric semipermeable barrier to ancient human migrations. Mol Biol Evol 29(1):359–365. https://doi.org/10.1093/molbev/msr221
Zytsar MV, Barashkov NA, Bady-Khoo MS, Shubina-Olejnik OA, Danilenko NG, Bondar AA, Morozov IV, Solovyev AV, Danilchenko VYu, Maximov VN, Posukh OL (2018) Updated carrier rates for c.35delG (GJB2) associated with hearing loss in Russia and common c.35delG haplotypes in Siberia. BMC Med Genet 19(1):138. https://doi.org/10.1186/s12881-018-0650-5
Zytsar MV, Bady-Khoo MS, Danilchenko VY, Maslova EA, Barashkov NA, Morozov IV, Bondar AA, Posukh OL (2020) High rates of three common GJB2 mutations c.516G>C, c.-23+1G>A, c.235delC in deaf patients from Southern Siberia are due to the founder effect. Genes (basel) 11(7):833. https://doi.org/10.3390/genes11070833
Acknowledgements
We thank all patients and blood sample donors who have contributed to this study. The authors are grateful to Anatoly N. Alekseev, Chandana Basu Mallik and Georgii P. Romanov for helpful comments on previous versions of this manuscript.
Funding
This study was supported by the Yakut Science Centre of Complex Medical Problems project: “Study of the genetic structure and burden of hereditary pathology of populations of the Republic of Sakha (Yakutia)”, the Ministry of Science and Higher Education of the Russian Federation (FSRG-2020-0016), the Russian Foundation for Basic Research (#18-05-60035_Arctika, # 20-015-00328_A, #19-34-60023_Perspektiva), the Russian State Budjet program No 0259-2021-0014, the European Union through the European Regional Development Fund (Project No. 2014-2020.4.01.15-0012) and IUT 24 Research Funding of the Institutional Estonian Ministry of Education and Research.
Author information
Authors and Affiliations
Contributions
Conceptualization: AVS, AK, NAB and SAF. Funding acquisition: SAF, AVS, VGP, OLP, and EM. Methodology: AVS and SAF. Project administration: SAF, NAB, MM, EKH, and AP. Resources: EB, M.B-Kh, MRL, TGM, GM, LK, and FMT. Visualization: AVS. Writing—original draft: AVS and NAB. Writing—review and editing: OLP, AK, and SAF.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical approval
This study was approved by the local Biomedical Ethics Committee at the Yakut Scientific Center of Complex Medical Problems, Yakutsk, Russia (Yakutsk, Protocol No. 50, 24 December 2019).
Consent to participate
Informed consent for genetic analyses was obtained from all individual participants included in this study.
Consent for publication
All the authors agreed that this manuscript be submitted to the journal of Human Genetics for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Solovyev, A.V., Kushniarevich, A., Bliznetz, E. et al. A common founder effect of the splice site variant c.-23 + 1G > A in GJB2 gene causing autosomal recessive deafness 1A (DFNB1A) in Eurasia. Hum Genet 141, 697–707 (2022). https://doi.org/10.1007/s00439-021-02405-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-021-02405-w