Abstract
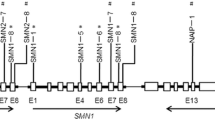
Spinal muscular atrophy (SMA) is a progressive motor neuron disease caused by loss or mutation of the survival motor neuron 1 (SMN1) gene and retention of SMN2. We performed targeted capture and sequencing of the SMN2, CFTR, and PLS3 genes in 217 SMA patients. We identified a 6.3 kilobase deletion that occurred in both SMN1 and SMN2 (SMN1/2) and removed exons 7 and 8. The deletion junction was flanked by a 21 bp repeat that occurred 15 times in the SMN1/2 gene. We screened for its presence in 466 individuals with the known SMN1 and SMN2 copy numbers. In individuals with 1 SMN1 and 0 SMN2 copies, the deletion occurred in 63% of cases. We modeled the deletion junction frequency and determined that the deletion occurred in both SMN1 and SMN2. We have identified the first deletion junction where the deletion removes exons 7 and 8 of SMN1/2. As it occurred in SMN1, it is a pathogenic mutation. We called variants in the PLS3 and SMN2 genes, and tested for association with mild or severe exception patients. The variants A-44G, A-549G, and C-1897T in intron 6 of SMN2 were significantly associated with mild exception patients, but no PLS3 variants correlated with severity. The variants occurred in 14 out of 58 of our mild exception patients, indicating that mild exception patients with an intact SMN2 gene and without modifying variants occur. This sample set can be used in the association analysis of candidate genes outside of SMN2 that modify the SMA phenotype.




Similar content being viewed by others
References
Ackermann B, Kröber S, Torres-Benito L et al (2013) Plastin 3 ameliorates spinal muscular atrophy via delayed axon pruning and improves neuromuscular junction functionality. Hum Mol Genet 1–20. https://doi.org/10.1093/hmg/dds540
Alvarado DM, Yang P, Druley TE et al (2014) Multiplexed direct genomic selection (MDiGS): a pooled BAC capture approach for highly accurate CNV and SNP/INDEL detection. Nucleic Acids Res 42:1–10. https://doi.org/10.1093/nar/gku218
Anhuf D, Eggermann T, Rudnik-Schöneborn S, Zerres K (2003) Determination of SMN1 and SMN2 copy number using TaqMan™ technology. Hum Mutat 22:74–78. https://doi.org/10.1002/humu.10221
Arkblad EL, Darin N, Berg K et al (2006) Multiplex ligation-dependent probe amplification improves diagnostics in spinal muscular atrophy. Neuromuscul Disord 16:830–838. https://doi.org/10.1016/j.nmd.2006.08.011
Arnold WD, Burghes AHM (2013) Spinal muscular atrophy: development and implementation of potential treatments. Ann Neurol 74:348–362. https://doi.org/10.1002/ana.23995
Arnold WD, Kassar D, Kissel JT (2015) Spinal muscular atrophy: diagnosis and management in a new therapeutic era. Muscle Nerve 51:157–167. https://doi.org/10.1002/mus.24497
Bernal S, Alías L, Barceló MJ et al (2010) The c.859G > C variant in the SMN2 gene is associated with types II and III SMA and originates from a common ancestor. J Med Genet 47:640–642. https://doi.org/10.1136/jmg.2010.079004
Bernal S, Also-Rallo E, Martínez-Hernández R et al (2011) Plastin 3 expression in discordant spinal muscular atrophy (SMA) siblings. Neuromuscul Disord 21:413–419. https://doi.org/10.1016/j.nmd.2011.03.009
Burghes AHM (1997) When is a deletion not a deletion? When it is converted. Am J Hum Genet 61:9–15. https://doi.org/10.1086/513913
Burghes AHM, Beattie CE (2009) Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci 10:597–609. https://doi.org/10.1038/nrn2670
Burghes AHM, McGovern VL (2017) Genetics of spinal muscular atrophy. Mol Cell Ther Mot Neuron Dis 121–139. https://doi.org/10.1016/B978-0-12-802257-3.00006-7
Burghes AHM, Ingraham SE, Kóte-Jarai Z et al (1994) Linkage mapping of the spinal muscular atrophy gene. Hum Genet 93:305–312
Burlet P, Bürglen L, Clermont O et al (1996) Large scale deletions of the 5q13 region are specific to Werdnig-Hoffmann disease. J Med Genet 33:281–283. https://doi.org/10.1136/jmg.33.4.281
Burnett BG, Muñoz E, Tandon A et al (2009) Regulation of SMN protein stability. Mol Cell Biol 29:1107–1115. https://doi.org/10.1128/MCB.01262-08
Calucho M, Bernal S, Alías L et al (2018) Correlation between SMA type and SMN2 copy number revisited: an analysis of 625 unrelated Spanish patients and a compilation of 2834 reported cases. Neuromuscul Disord 28:208–215. https://doi.org/10.1016/j.nmd.2018.01.003
Campbell L, Potter A, Ignatius J et al (1997) Genomic variation and gene conversion in spinal muscular atrophy: implications for disease process and clinical phenotype. Am J Hum Genet 61:40–50. https://doi.org/10.1086/513886
Carpten JD, DiDonato CJ, Ingraham SE et al (1994) A YAC contig of the region containing the spinal muscular atrophy gene (SMA): identification of an unstable region. Genomics 24:351–356. https://doi.org/10.1006/geno.1994.1626
Cartegni L, Krainer AR (2002) Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN. Nat Genet 30:377–384. https://doi.org/10.1038/ng854
Ceballos FC, Hazelhurst S, Ramsay M (2018) Assessing runs of Homozygosity: a comparison of SNP Array and whole genome sequence low coverage data. BMC Genom 19:1–12. https://doi.org/10.1186/s12864-018-4489-0
Chen Q, Baird SD, Mahadevan M et al (1998) Sequence of a 131-kb region of 5q13.1 containing the spinal muscular atrophy candidate genes SMN and NAIP. Genomics 48:121–127. https://doi.org/10.1006/geno.1997.5141
Cobben JM, Van Der Steege G, Grootscholten P et al (1995) Deletions of the survival motor neuron gene in unaffected siblings of patients with spinal muscular atrophy. Am J Hum Genet 57:805–808
Coovert DD, Le TT, McAndrew PE et al (1997) The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet 6:1205–1214. https://doi.org/10.1093/hmg/6.8.1205
Crawford TO, Pardo CA (1996) The neurobiology of childhood spinal muscular atrophy. Neurobiol Dis 3:97–110. https://doi.org/10.1006/nbdi.1996.0010
Cuscó I, Barceló MJ, Rojas-García R et al (2006) SMN2 copy number predicts acute or chronic spinal muscular atrophy but does not account for intrafamilial variability in siblings. J Neurol 253:21–25. https://doi.org/10.1007/s00415-005-0912-y
DePristo M, Banks E, Poplin R et al (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. https://doi.org/10.1038/ng.806
DiDonato CJ (1995) The spinal muscular atrophy gene: “isolation and characterization of the genetic and physical region surrounding the gene locus and identification of candidate cDNAs. The Ohio State University, Columbus, OH
DiDonato CJ, Morgan K, Carpten JD et al (1994) Association between Ag1-CA alleles and severity of autosomal recessive proximal spinal muscular atrophy. Am J Hum Genet 55:1218–1229
DiDonato CJ, Ingraham SE, Mendell JR et al (1997) Deletion and conversion in spinal muscular atrophy patients: is there a relationship to severity? Ann Neurol 41:230–237. https://doi.org/10.1002/ana.410410214
Dobin A, Davis CA, Schlesinger F et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. https://doi.org/10.1093/bioinformatics/bts635
Eisfeldt J, Nilsson D, Andersson-Assarsson JC, Lindstrand A (2018) AMYCNE: Confident copy number assessment using whole genome sequencing data. PLoS One 13:1–14. https://doi.org/10.1371/journal.pone.0189710
Feldkötter M, Schwarzer V, Wirth R et al (2002) Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet 70:358–368. https://doi.org/10.1086/338627
Finkel RS, Mercuri E, Darras BT et al (2017) Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N Engl J Med 377:1723–1732. https://doi.org/10.1056/NEJMoa1702752
Gidaro T, Servais L (2018) Nusinersen treatment of spinal muscular atrophy: current knowledge and existing gaps. Dev Med Child Neurol. https://doi.org/10.1111/dmcn.14027
Hahnen E, Forkert R, Marke C et al (1995) Molecular analysis of candidate genes on chromosome 5q13 in autosomal recessive spinal muscular atrophy: evidence of homozygous deletions of the SMN gene in unaffected individuals. Hum Mol Genet 4:1927–1933. https://doi.org/10.1093/hmg/4.10.1927
Hao LT, Burghes AHM, Beattie CE (2011) Generation and characterization of a genetic zebrafish model of SMA carrying the human SMN2 gene. Mol Neurodegener 6:1–9. https://doi.org/10.1186/1750-1326-6-24
Hauke J, Riessland M, Lunke S et al (2009) Survival motor neuron gene 2 silencing by DNA methylation correlates with spinal muscular atrophy disease severity and can be bypassed by histone deacetylase inhibition. Hum Mol Genet 18:304–317. https://doi.org/10.1093/hmg/ddn357
Jedrzejowska M, Borkowska J, Zimowski J et al (2008) Unaffected patients with a homozygous absence of the SMN1 gene. Eur J Hum Genet 16:930–934. https://doi.org/10.1038/ejhg.2008.41
Jedrzejowska M, Milewski M, Zimowski J et al (2009) Phenotype modifiers of spinal muscular atrophy: the number of SMN2 gene copies, deletion in the NAIP gene and probably gender influence the course of the disease. Acta Biochim Pol 56:103–108
Kashima T, Manley JL (2003) A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet 34:460–463. https://doi.org/10.1038/ng1207
Le TT, Pham LT, Butchbach MER et al (2005) SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet 14:845–857. https://doi.org/10.1093/hmg/ddi078
Lefebvre S, Bürglen L, Reboullet S et al (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80:155–165
Lefebvre S, Burlet P, Liu Q et al (1997) Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet 16:265–269. https://doi.org/10.1038/ng0797-265
Li H, Handsaker B, Wysoker A et al (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Lorson CL, Strasswimmer J, Yao JM et al (1998) SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet 19:63–66. https://doi.org/10.1038/ng0598-63
Lorson CL, Hahnen E, Androphy EJ, Wirth B (1999) A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA 96:6307–6311. https://doi.org/10.1073/pnas.96.11.6307
Mailman MD, Heinz JW, Papp AC et al (2002) Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet Med 4:20–26. https://doi.org/10.1097/00125817-200201000-00004
McAndrew PE, Parsons DW, Simard LR et al (1997) Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet 60:1411–1422. https://doi.org/10.1086/515465
McGovern VL, Massoni-Laporte A, Wang X et al (2015) Plastin 3 expression does not modify spinal muscular atrophy severity in the ∆7 SMA mouse. PLoS One 10:1–19. https://doi.org/10.1371/journal.pone.0132364
Melki J, Lefebvre S, Burglen L et al (1994) De novo and inherited deletions of the 5q13 region in spinal muscular atrophies. Science 264:1474–1477. https://doi.org/10.1126/science.7910982
Mendell JR, Al-Zaidy S, Shell R et al (2017) Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med 377:1713–1722. https://doi.org/10.1056/NEJMoa1706198
Mercuri E, Darras BT, Chiriboga CA et al (2018) Nusinersen versus sham control in later-onset spinal muscular atrophy. N Engl J Med 378:625–635. https://doi.org/10.1056/NEJMoa1710504
Miller RG, Moore DH, Dronsky V et al (2001) A placebo-controlled trial of gabapentin in spinal muscular atrophy. J Neurol Sci 191:127–131. https://doi.org/10.1016/S0022-510X(01)00632-3
Monani UR, Lorson CL, Parsons DW et al (1999) A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet 8:1177–1183. https://doi.org/10.1093/hmg/8.7.1177
Munsat TL (1991) International SMA Collaboration. Neuromuscul Disord 1:81. https://doi.org/10.1016/0960-8966(91)90052-T
Oprea GE, Kröber S, McWhorter ML et al (2008) Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science 320:524–527. https://doi.org/10.1126/science.1155085
Osoegawa K, Woon PY, Zhao B et al (1998) An improved approach for construction of bacterial artificial chromosome libraries. Genomics 52:1–8. https://doi.org/10.1006/geno.1998.5423
Osoegawa K, Mammoser AG, Wu C et al (2001) A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res 11:483–496. https://doi.org/10.1101/gr.169601
Pane M, Lapenta L, Abiusi E et al (2017) Longitudinal assessments in discordant twins with SMA. Neuromuscul Disord 27:890–893. https://doi.org/10.1016/j.nmd.2017.06.559
Pearn JH (1973) The gene frequency of acute werdnig-hoffmann disease (SMA type 1). A total population survey in North-East England. J Med Genet 10:260–265. https://doi.org/10.1136/jmg.10.3.260
Pearn JH (1978) Incidence, prevalence, and gene frequency studies of chronic childhood spinal muscular atrophy. J Med Genet 15:409–413. https://doi.org/10.1136/jmg.15.6.409
Prior TW (2007) Spinal muscular atrophy diagnostics. J Child Neurol 22:952–956. https://doi.org/10.1177/0883073807305668
Prior TW, Swoboda KJ, Scott HD, Hejmanowski AQ (2004) Homozygous SMN1 deletions in unaffected family members and modification of the phenotype by SMN2. Am J Med Genet 130 A:307–310. https://doi.org/10.1002/ajmg.a.30251
Prior TW, Krainer AR, Hua Y et al (2009) A positive modifier of spinal muscular atrophy in the SMN2 gene. Am J Hum Genet 85:408–413. https://doi.org/10.1016/j.ajhg.2009.08.002
Pyatt RE, Prior TW (2006) A feasibility study for the newborn screening of spinal muscular atrophy. Genet Med 8:428–437. https://doi.org/10.1097/01.gim.0000227970.60450.b2
R Core Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Riessland M, Kaczmarek A, Schneider S et al (2017) Neurocalcin delta suppression protects against spinal muscular atrophy in humans and across species by restoring impaired endocytosis. Am J Hum Genet 100:297–315. https://doi.org/10.1016/j.ajhg.2017.01.005
Roberts DF, Chavez J, Court SDM (1970) The genetic component in child mortality. Arch Dis Child 45:33–38. https://doi.org/10.1136/adc.45.239.33
Roy N, Mahadevan MS, McLean M et al (1995) The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell 80:167–178. https://doi.org/10.1016/0092-8674(95)90461-1
Rüdiger NS, Gregersen N, Kielland-brandt MC (1995) One short well conserved region of Alu-sequences is involved in human gene rearrangements and has homology with prokaryotic chi. Nucleic Acids Res 23:256–260. https://doi.org/10.1093/nar/23.2.256
Strathmann EA, Peters M, Hosseinibarkooie S et al (2018) Evaluation of potential effects of Plastin 3 overexpression and low-dose SMN-antisense oligonucleotides on putative biomarkers in spinal muscular atrophy mice. PLoS One 4:1–28
Sugarman EA, Nagan N, Zhu H et al (2012) Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of 472400 specimens. Eur J Hum Genet 20:27–32. https://doi.org/10.1038/ejhg.2011.134
Szkandera J, Winder T, Stotz M et al (2013) A common gene variant in PLS3 predicts colon cancer recurrence in women. Tumor Biol 34:2183–2188. https://doi.org/10.1007/s13277-013-0754-7
Thompson TG, Didonato CJ, Simard LR et al (1995) A novel cDNA detects homozygous microdeletions in greater than 50% of type I spinal muscular atrophy patients. Nat Genet 9:56–62. https://doi.org/10.1038/ng0195-56
Velasco E, Valero C, Valero A et al (1996) Molecular analysis of the SMN and NAIP genes in Spanish spinal muscular atrophy (SMA) families and correlation between number of copies ofcBCD541 and SMA phenotype. Hum Mol Genet 5:257–263. https://doi.org/10.1093/hmg/5.2.257
Vezain M, Saugier-Veber P, Goina E et al (2010) A rare SMN2 variant in a previously unrecognized composite splicing regulatory element induces exon 7 inclusion and reduces the clinical severity of spinal muscular atrophy. Hum Mutat 31:1110–1125. https://doi.org/10.1002/humu.21173
Wang CC, Chang JG, Chen YL et al (2010) Multi-exon genotyping of SMN gene in spinal muscular atrophy by universal fluorescent PCR and capillary electrophoresis. Electrophoresis 31:2396–2404. https://doi.org/10.1002/elps.201000124
Weber JL, Polymeropoulos MH, May PE et al (1991) Mapping of human chromosome 5 microsatellite DNA polymorphisms. Genomics 11:695–700. https://doi.org/10.1016/0888-7543(91)90077-R
Wirth B, El-Agwany A, Baasner A et al (1995) Mapping of the spinal muscular atrophy (SMA) gene to a 750-kb interval flanked by two new microsatellites. Eur J Hum Genet 3:56–60. https://doi.org/10.1159/000472274
Wirth B, Schmidt T, Hahnen E et al (1997) De novo rearrangements found in 2% of index patients with spinal muscular atrophy: mutational mechanisms, parental origin, mutation rate, and implications for genetic counseling. Am J Hum Genet 61:1102–1111. https://doi.org/10.1086/301608
Wirth B, Herz M, Wetter A et al (1999) Quantitative analysis of survival motor neuron copies: identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype-phenotype correlation, and implications for genetic counseling. Am J Hum Genet 64:1340–1356
Wu X, Wang S-H, Sun J et al (2017) A-44G transition in SMN2 intron 6 protects patients with spinal muscular atrophy. Hum Mol Genet 26:2768–2780. https://doi.org/10.1093/hmg/ddx166
Zerres K, Rudnik-Schoneborn S (1995) Natural history in proximal spinal muscular atrophy: clinical analysis of 445 patients and suggestions for a modification of existing classifications. Arch Neurol 52:518–523
Zheleznyakova GY, Voisin S, Kiselev AV et al (2013) Genome-wide analysis shows association of epigenetic changes in regulators of Rab and Rho GTPases with spinal muscular atrophy severity. Eur J Hum Genet 21:988–993. https://doi.org/10.1038/ejhg.2012.293
Zheleznyakova GY, Nilsson EK, Kiselev AV et al (2015) Methylation levels of SLC23A2 and NCOR2 genes correlate with spinal muscular atrophy severity. PLoS One 10:1–14. https://doi.org/10.1371/journal.pone.0121964
Acknowledgements
These studies were supported by a grant from the Muscular Dystrophy Association MDA352913. In addition, funding was provided from the National Institutes of Child Health and Development (NICHD) R01HD069045 Award to KJS for development of the Project Cure SMA Longitudinal Pediatric Data Repository. Corey Ruhno was supported by NICHD grant HD060586. We would also like to thank the Ohio Supercomputing Center. Anton J. Blatnik III assisted with sequencing of polymorphic markers. Finally, thank you to Dr. Jennifer Sinnott in The Ohio State University Department of Statistics for assisting us with the development of the models used in this paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ruhno, C., McGovern, V.L., Avenarius, M.R. et al. Complete sequencing of the SMN2 gene in SMA patients detects SMN gene deletion junctions and variants in SMN2 that modify the SMA phenotype. Hum Genet 138, 241–256 (2019). https://doi.org/10.1007/s00439-019-01983-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-019-01983-0