Abstract
Long non-coding RNAs (lncRNAs) serve as important controller of cellular functions via regulating RNA transcription, degradation and translation. However, what are the regulation patterns of lncRNAs on downstream mRNA and how the upstream genetic variants regulate lncRNAs are largely unknown. We first performed a comprehensive expression quantitative trait locus (eQTL) analysis (MatrixeQTL package, R) using genome-wide lncRNA expression and SNP genotype data from human peripheral blood mononuclear cells (PBMCs) of 43 unrelated individuals. Subsequently, multi-omics integrative network analysis was applied to construct SNP-lncRNA-mRNA (SLM) interaction networks. The causal inference test (CIT) was used to identify lncRNA-mediated (epi-) genetic regulation on mRNA expressions. Our eQTL analysis detected 707 pairs of cis-effect associations (p < 5.64E−06) and 6657 trans-effect associations (p < 3.51E−08), respectively. We also found that top significant cis-eSNPs were enriched around the lncRNA transcription start site regions, and that enrichment patterns of cis-eSNPs differs among different lncRNA sizes (small, medium and large).The constructed SLM interaction networks (1 primary networks and four small separate networks) showed various complex interaction patterns. Especially, the in-depth CIT detected 50 significant lncRNA-mediated SLM trios, and some hotspots (e.g., SNPs: rs926370, rs7716167 and rs16880521; lncRNAs: HIT000061975 and ENST00000579057.1). This study represents the first effort of dissecting the SLM interaction patterns in PBMCs by multi-omics integrative network analysis and causal inference test for clearing the regulation chain. The results provide novel insights into the regulation patterns of lncRNA, and may facilitate investigations of PBMC-related immune physiological process and immunological diseases in the future.






Similar content being viewed by others
Abbreviations
- 3′UTRs:
-
3′ Untranslated regions
- CIT:
-
Causal inference test
- DAVID:
-
The database for annotation, visualization, and integrated discovery
- elncRNA:
-
eQTL lncRNA
- eProbe:
-
eQTL probe
- eQTL:
-
Expression quantitative trait locus
- eSNP:
-
eQTL SNPs
- FDR:
-
False-discovery rate
- GO:
-
Gene ontology
- GWAS:
-
Genome-wide association studies
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- LD:
-
Linkage disequilibrium
- lncR-eQTLs:
-
lncRNA eQTLs
- lncRNAs:
-
Long non-coding RNAs
- PBMCs:
-
Peripheral blood mononuclear cells
- PCC:
-
Pearson correlation coefficients
- RA:
-
Rheumatoid arthritis
- SLM:
-
SNP-lncRNA-mRNA
- SNPs:
-
Single nucleotide polymorphisms
- TES:
-
Transcription end site
- TFBS:
-
Transcription factor binding sites
- TSS:
-
Transcription start site
References
Abecasis GR et al (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. doi:10.1038/nature11632
Adzhubei IA et al (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249. doi:10.1038/nmeth0410-248
Altshuler D, Daly MJ, Lander ES (2008) Genetic mapping in human disease. Science 322:881–888. doi:10.1126/science.1156409
Borel C et al (2011) Identification of cis- and trans-regulatory variation modulating microRNA expression levels in human fibroblasts. Genome Res 21:68–73. doi:10.1101/gr.109371.110
Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, Gabellini D (2012) A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell 149:819–831. doi:10.1016/j.cell.2012.03.035
Derrien T et al (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22:1775–1789. doi:10.1101/gr.132159.111
Dimas AS et al (2009) Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325:1246–1250. doi:10.1126/science.1174148
Gerstein MB et al (2012) Architecture of the human regulatory network derived from ENCODE data. Nature 489:91–100. doi:10.1038/nature11245
Gupta RA et al (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464:1071–1076. doi:10.1038/nature08975
Guttman M et al (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458:223–227. doi:10.1038/nature07672
Huan T et al (2015) Genome-wide identification of microRNA expression quantitative trait loci Nat Commun 6:6601. doi:10.1038/ncomms7601
Huang T, Cai YD (2013) An information-theoretic machine learning approach to expression QTL analysis. PLoS One 8:e67899. doi:10.1371/journal.pone.0067899
Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi:10.1038/nprot.2008.211
Huarte M, Rinn JL (2010) Large non-coding RNAs: missing links in cancer? Hum Mol Genet 19:R152–R161. doi:10.1093/hmg/ddq353
Hubbard T et al (2002) The Ensembl genome database project. Nucleic Acids Res 30:38–41
Kellis M et al (2014) Defining functional DNA elements in the human genome. Proc Natl Acad Sci USA 111:6131–6138. doi:10.1073/pnas.1318948111
Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP (2010) Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 3:ra8. doi:10.1126/scisignal.2000568
Klattenhoff CA et al (2013) Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152:570–583. doi:10.1016/j.cell.2013.01.003
Kumar V, Wijmenga C, Withoff S (2012) From genome-wide association studies to disease mechanisms: celiac disease as a model for autoimmune diseases. Semin Immunopathol 34:567–580. doi:10.1007/s00281-012-0312-1
Kumar V et al (2013) Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet 9:e1003201. doi:10.1371/journal.pgen.1003201
Lettre G, Rioux JD (2008) Autoimmune diseases: insights from genome-wide association studies. Hum Mol Genet 17:R116–R121. doi:10.1093/hmg/ddn246
Liu Y et al (2013) Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol 31:142–147. doi:10.1038/nbt.2487
McCarthy MI, Hirschhorn JN (2008) Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet 17:R156–R165. doi:10.1093/hmg/ddn289
Meola N, Pizzo M, Alfano G, Surace EM, Banfi S (2012) The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. RNA 18:111–123. doi:10.1261/rna.029454.111
Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10:155–159. doi:10.1038/nrg2521
Millstein J, Zhang B, Zhu J, Schadt EE (2009) Disentangling molecular relationships with a causal inference test. BMC Genet 10:23. doi:10.1186/1471-2156-10-23
Murphy A et al (2010) Mapping of numerous disease-associated expression polymorphisms in primary peripheral blood CD4+ lymphocytes. Hum Mol Genet 19:4745–4757. doi:10.1093/hmg/ddq392
Niazi F, Valadkhan S (2012) Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3′ UTRs. RNA 18:825-843. doi:10.1261/rna.029520.111
Pennisi E (2010) Genomics. 1000 Genomes Project gives new map of genetic diversity. Science 330:574–575. doi:10.1126/science.330.6004.574
Pennisi E (2011) The biology of genomes. Disease risk links to gene regulation. Science 332:1031. doi:10.1126/science.332.6033.1031
Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136:629–641. doi:10.1016/j.cell.2009.02.006
Prensner JR, Chinnaiyan AM (2011) The emergence of lncRNAs in cancer biology. Cancer Discov 1:391–407. doi:10.1158/2159-8290.CD-11-0209
Qi J, Asl HF, Bjorkegren J, Michoel T (2014) kruX: matrix-based non-parametric eQTL discovery. BMC Bioinform 15:11. doi:10.1186/1471-2105-15-11
Schadt EE et al (2003) Genetics of gene expression surveyed in maize, mouse and man. Nature 422:297–302. doi:10.1038/nature01434
Schonrock N, Harvey RP, Mattick JS (2012) Long noncoding RNAs in cardiac development and pathophysiology. Circ Res 111:1349–1362. doi:10.1161/CIRCRESAHA.112.268953
Shabalin AA (2012) Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28:1353–1358. doi:10.1093/bioinformatics/bts163
Spizzo R, Almeida MI, Colombatti A, Calin GA (2012) Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene 31:4577–4587. doi:10.1038/onc.2011.621
Tripathi V et al (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39:925–938. doi:10.1016/j.molcel.2010.08.011
van Riel PL, Fransen J (2005) DAS28: a useful instrument to monitor infliximab treatment in patients with rheumatoid arthritis. Arthritis Res Ther 7:189–190. doi:10.1186/ar1820
Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, Pritchard JK (2008) High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet 4:e1000214. doi:10.1371/journal.pgen.1000214
Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43:904–914. doi:10.1016/j.molcel.2011.08.018
Wen X, Luca F, Pique-Regi R (2015) Cross-population joint analysis of eQTLs: fine mapping and functional annotation. PLoS Genet 11:e1005176. doi:10.1371/journal.pgen.1005176
Westra HJ et al (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45:1238–1243. doi:10.1038/ng.2756
Willingham AT et al (2005) A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309:1570–1573. doi:10.1126/science.1115901
Xu Z, Taylor JA (2009) SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res 37:W600–W605. doi:10.1093/nar/gkp290
Yue P, Melamud E, Moult J (2006) SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinform 7:166. doi:10.1186/1471-2105-7-166
Acknowledgements
The study was supported by Natural Science Foundation of China (81473046, 31271336, 81373010, 81502868, 31401079, 81401343, 81541068), the Natural Science Research Project of Jiangsu Provincial Higher Education (16KJA330001), the Natural Science Foundation of Jiangsu Province (BK20130300, BK20150346), the Project funded by China PostdoctoralScience Foundation (2014M551649),the Startup Fund from Soochow University (Q413900112, Q413900712), and a Project of the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
439_2017_1771_MOESM2_ESM.tif
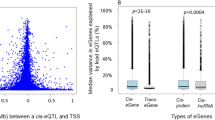
Supplementary material 2 (TIFF 164 kb) Figure S1. Distribution of eQTL association significance (−log10 p value) for eSNPs against their distance (bp) from TSS in different types of lncRNA
Rights and permissions
About this article
Cite this article
Xia, W., Zhu, XW., Mo, XB. et al. Integrative multi-omics analysis revealed SNP-lncRNA-mRNA (SLM) networks in human peripheral blood mononuclear cells. Hum Genet 136, 451–462 (2017). https://doi.org/10.1007/s00439-017-1771-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-017-1771-1