Abstract
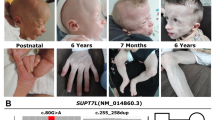
Impairment of ubiquitin–proteasome system activity involving ubiquitin ligase genes UBE3A, UBE3B, and HUWE1 and deubiquitinating enzyme genes USP7 and USP9X has been reported in patients with neurodevelopmental delays. To date, only a handful of single-nucleotide variants (SNVs) and copy-number variants (CNVs) involving TRIP12, encoding a member of the HECT domain E3 ubiquitin ligases family on chromosome 2q36.3 have been reported. Using chromosomal microarray analysis and whole-exome sequencing (WES), we have identified, respectively, five deletion CNVs and four inactivating SNVs (two frameshifts, one missense, and one splicing) in TRIP12. Seven of these variants were found to be de novo; parental studies could not be completed in two families. Quantitative PCR analyses of the splicing mutation showed a dramatically decreased level of TRIP12 mRNA in the proband compared to the family controls, indicating a loss-of-function mechanism. The shared clinical features include intellectual disability with or without autistic spectrum disorders, speech delay, and facial dysmorphism. Our findings demonstrate that E3 ubiquitin ligase TRIP12 plays an important role in nervous system development and function. The nine presented pathogenic variants further document that TRIP12 haploinsufficiency causes a childhood-onset neurodevelopmental disorder. Finally, our data enable expansion of the phenotypic spectrum of ubiquitin–proteasome dependent disorders.



Similar content being viewed by others
References
Aravind L (2001) The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem Sci 26:273–275
Boone PM, Bacino CA, Shaw CA, Eng PA, Hixson PM, Pursley AN, Kang SH, Yang Y, Wiszniewska J, Nowakowska BA, del Gaudio D, Xia Z, Simpson-Patel G, Immken LL, Gibson JB, Tsai AC, Bowers JA, Reimschisel TE, Schaaf CP, Potocki L, Scaglia F, Gambin T, Sykulski M, Bartnik M, Derwinska K, Wisniowiecka-Kowalnik B, Lalani SR, Probst FJ, Bi W, Beaudet AL, Patel A, Lupski JR, Cheung SW, Stankiewicz P (2010) Detection of clinically relevant exonic copy-number changes by array CGH. Hum Mutat 31:1326–1342
Bramswig NC, Lüdecke HJ, Pettersson M, Albrecht B, Bernier RA, Cremer K, Eichler EE, Falkenstein D, Gerdts J, Jansen S, Kuechler A, Kvarnung M, Lindstrand A, Nilsson D, Nordgren A, Pfundt R, Spruijt L, Surowy HM, de Vries BB, Wieland T, Engels H, Strom TM, Kleefstra T, Wieczorek D (2017) Identification of new TRIP12 variants and detailed clinical evaluation of individuals with non-syndromic intellectual disability with or without autism. Hum Genet 136(2):179–192
Chen D, Kon N, Zhong J, Zhang P, Yu L, Gu W (2013) Differential effects on ARF stability by normal versus oncogenic levels of c-Myc expression. Mol Cell 51:46–56
Chen D, Shan J, Zhu WG, Qin J, Gu W (2010) Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature 464:624–627
Chen Y, McPhie DL, Hirschberg J, Neve RL (2000) The amyloid precursor protein-binding protein APP-BP1 drives the cell cycle through the S-M checkpoint and causes apoptosis in neurons. J Biol Chem 275:8929–8935
Doco-Fenzy M, Landais E, Andrieux J, Schneider A, Delemer B, Sulmont V, Melin JP, Ploton D, Thevenard J, Monboisse JC, Belouadah M, Lefebvre F, Durlach A, Goossens M, Albuisson J, Motte J, Gaillard D (2008) Deletion 2q36.2q36.3 with multiple renal cysts and severe mental retardation. Eur J Med Genet 51:598–607
Gudjonsson T, Altmeyer M, Savic V, Toledo L, Dinant C, Grøfte M, Bartkova J, Poulsen M, Oka Y, Bekker-Jensen S, Mailand N, Neumann B, Heriche JK, Shearer R, Saunders D, Bartek J, Lukas J, Lukas C (2012) TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell 150:697–709
Hanoun N, Fritsch S, Gayet O, Gigoux V, Cordelier P, Dusetti N, Torrisani J, Dufresne M (2014) The E3 ubiquitin ligase thyroid hormone receptor-interacting protein 12 targets pancreas transcription factor 1a for proteasomal degradation. J Biol Chem 289:35593–35604
Kajiro M, Tsuchiya M, Kawabe Y, Furumai R, Iwasaki N, Hayashi Y et al (2011) The E3 ubiquitin ligase activity of Trip12 is essential for mouse embryogenesis. PLoS ONE 6:e25871
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Exome Aggregation Consortium (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285–291
Lelieveld SH, Reijnders MR, Pfundt R, Yntema HG, Kamsteeg EJ, de Vries P, de Vries BB, Willemsen MH, Kleefstra T, Löhner K, Vreeburg M, Stevens SJ, van der Burgt I, Bongers EM, Stegmann AP, Rump P, Rinne T, Nelen MR, Veltman JA, Vissers LE, Brunner HG, Gilissen C (2016) Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci 19:1194–1196
Oikonomakis V, Kosma K, Mitrakos A, Sofocleous C, Pervanidou P, Syrmou A, Pampanos A, Psoni S, Fryssira H, Kanavakis E, Kitsiou-Tzeli S, Tzetis M (2016) Recurrent copy number variations as risk factors for autism spectrum disorders: analysis of the clinical implications. Clin Genet 89:708–718
O’Roak BJ, Stessman HA, Boyle EA, Witherspoon KT, Martin B, Lee C, Vives L, Baker C, Hiatt JB, Nickerson DA, Bernier R, Shendure J, Eichler EE (2014) Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun 5:5595
Park Y, Yoon SK, Yoon JB (2008) TRIP12 functions as an E3 ubiquitin ligase of APP-BP1. Biochem Biophys Res Commun 374:294–298
Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, Thiruvahindrapuram B, Xu X, Ziman R, Wang Z, Vorstman JA, Thompson A, Regan R, Pilorge M, Pellecchia G, Pagnamenta AT, Oliveira B, Marshall CR, Magalhaes TR, Lowe JK, Howe JL, Griswold AJ, Gilbert J, Duketis E, Dombroski BA, De Jonge MV, Cuccaro M, Crawford EL, Correia CT, Conroy J, Conceição IC, Chiocchetti AG, Casey JP, Cai G, Cabrol C, Bolshakova N, Bacchelli E, Anney R, Gallinger S, Cotterchio M, Casey G, Zwaigenbaum L, Wittemeyer K, Wing K, Wallace S, van Engeland H, Tryfon A, Thomson S, Soorya L, Rogé B, Roberts W, Poustka F, Mouga S, Minshew N, McInnes LA, McGrew SG, Lord C, Leboyer M, Le Couteur AS, Kolevzon A, Jiménez González P, Jacob S, Holt R, Guter S, Green J, Green A, Gillberg C, Fernandez BA, Duque F, Delorme R, Dawson G, Chaste P, Café C, Brennan S, Bourgeron T, Bolton PF, Bölte S, Bernier R, Baird G, Bailey AJ, Anagnostou E, Almeida J, Wijsman EM, Vieland VJ, Vicente AM, Schellenberg GD, Pericak-Vance M, Paterson AD, Parr JR, Oliveira G, Nurnberger JI, Monaco AP, Maestrini E, Klauck SM, Hakonarson H, Haines JL, Geschwind DH, Freitag CM, Folstein SE, Ennis S, Coon H, Battaglia A, Szatmari P, Sutcliffe JS, Hallmayer J, Gill M, Cook EH, Buxbaum JD, Devlin B, Gallagher L, Betancur C, Scherer SW (2014) Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet 94:677–694
Poulsen EG, Steinhauer C, Lees M, Lauridsen AM, Ellgaard L, Hartmann-Petersen R (2012) HUWE1 and TRIP12 collaborate in degradation of ubiquitin-fusion proteins and misframed ubiquitin. PLoS ONE 7:e50548
Rotin D, Kumar S (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 10:398–409
Velimezi G, Liontos M, Vougas K, Roumeliotis T, Bartkova J, Sideridou M, Dereli-Oz A, Kocylowski M, Pateras IS, Evangelou K, Kotsinas A, Orsolic I, Bursac S, Cokaric-Brdovcak M, Zoumpourlis V, Kletsas D, Papafotiou G, Klinakis A, Volarevic S, Gu W, Bartek J, Halazonetis TD, Gorgoulis VG (2013) Functional interplay between the DNA-damage-response kinase ATM and ARF tumour suppressor protein in human cancer. Nat Cell Biol 15:967–977
Wiszniewska J, Bi W, Shaw C, Stankiewicz P, Kang SH, Pursley AN, Lalani S, Hixson P, Gambin T, Tsai CH, Bock HG, Descartes M, Probst FJ, Scaglia F, Beaudet AL, Lupski JR, Eng C, Cheung SW, Bacino C, Patel A (2014) Combined array CGH plus SNP genome analyses in a single assay for optimized clinical testing. Eur J Hum Genet 22:79–87
Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, Veeraraghavan N, Hawes A, Chiang T, Leduc M, Beuten J, Zhang J, He W, Scull J, Willis A, Landsverk M, Craigen WJ, Bekheirnia MR, Stray-Pedersen A, Liu P, Wen S, Alcaraz W, Cui H, Walkiewicz M, Reid J, Bainbridge M, Patel A, Boerwinkle E, Beaudet AL, Lupski JR, Plon SE, Gibbs RA, Eng CM (2014) Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 312:1870–1879
URLs
1000 Genomes Project. http://www.1000genomes.org. Accessed 11 Oct 2016
Atherosclerosis Risk in Communities Study. http://www2.cscc.unc.edu/aric. Accessed 11 Oct 2016
ExAC Brower. Exome Aggregation Consortium. http://exac.broadinstitute.org. Accessed 11 Oct 2016
Mutation Taster. http://www.mutationtaster.org. Accessed 11 Oct 2016
NHLBI Exome Sequencing Project (ESP). Exome Variant Server. http://evs.gs.washington.edu/EVS. Accessed 11 Oct 2016
Online Mendelian Inheritance in Man (OMIM). http://www.omim.org. Accessed 11 Oct 2016
PolyPhen-2. http://genetics.bwh.harvard.edu/pph2. Accessed 11 Oct 2016
Acknowledgements
We are grateful to the families for participating in this study. We are most grateful to the Genomics platform of Nantes (Biogenouest Genomics) core facility for its technical support on performing exome for Subject 9 and his parents. We acknowledge HUGODIMS (Western France exome-based trio approach project to identify genes involved in intellectual disability). This study makes use of data generated by the DECIPHER community. A full list of centres who contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the project was provided by the Wellcome Trust. This work was supported in part by the National Human Genome Research Institute/National Heart Lung and Blood Institute grant to the Baylor-Hopkins Center for Mendelian Genomics [U54HG006542], and the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under Award Number U01HG007709, the National Institute of Neurological Disorders and Stroke R01 Grant [NS058529 to J.R.L.], and a grant from the French Ministry of Health and Poitou–Charentes Regional Health Agency (HUGODIMS, 2013, RC14_0107).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
JRL has stock ownership in 23andMe and Lasergen, is a paid consultant for Regeneron Pharmaceuticals, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis and clinical exome sequencing offered by Baylor Genetics (http://bmgl.com/).
Additional information
J. Zhang and T. Gambin contributed equally to this work.
An erratum to this article is available at http://dx.doi.org/10.1007/s00439-017-1828-1.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, J., Gambin, T., Yuan, B. et al. Haploinsufficiency of the E3 ubiquitin-protein ligase gene TRIP12 causes intellectual disability with or without autism spectrum disorders, speech delay, and dysmorphic features. Hum Genet 136, 377–386 (2017). https://doi.org/10.1007/s00439-017-1763-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-017-1763-1