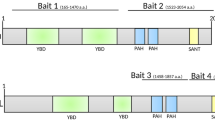
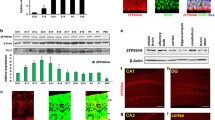
Abstract
GTF2IRD1 is one of the three members of the GTF2I gene family, clustered on chromosome 7 within a 1.8 Mb region that is prone to duplications and deletions in humans. Hemizygous deletions cause Williams–Beuren syndrome (WBS) and duplications cause WBS duplication syndrome. These copy number variations disturb a variety of developmental systems and neurological functions. Human mapping data and analyses of knockout mice show that GTF2IRD1 and GTF2I underpin the craniofacial abnormalities, mental retardation, visuospatial deficits and hypersociability of WBS. However, the cellular role of the GTF2IRD1 protein is poorly understood due to its very low abundance and a paucity of reagents. Here, for the first time, we show that endogenous GTF2IRD1 has a punctate pattern in the nuclei of cultured human cell lines and neurons. To probe the functional relationships of GTF2IRD1 in an unbiased manner, yeast two-hybrid libraries were screened, isolating 38 novel interaction partners, which were validated in mammalian cell lines. These relationships illustrate GTF2IRD1 function, as the isolated partners are mostly involved in chromatin modification and transcriptional regulation, whilst others indicate an unexpected role in connection with the primary cilium. Mapping of the sites of protein interaction also indicates key features regarding the evolution of the GTF2IRD1 protein. These data provide a visual and molecular basis for GTF2IRD1 nuclear function that will lead to an understanding of its role in brain, behaviour and human disease.








Similar content being viewed by others
Abbreviations
- hESC:
-
Human embryonic stem cells
- PLA:
-
Proximity ligation assay
- STED:
-
Stimulated emission depletion
- WBS:
-
Williams–Beuren syndrome
- Y2H:
-
Yeast two-hybrid
References
Antonell A, Del Campo M, Magano LF, Kaufmann L, de la Iglesia JM, Gallastegui F, Flores R, Schweigmann U, Fauth C, Kotzot D, Perez-Jurado LA (2010) Partial 7q11.23 deletions further implicate GTF2I and GTF2IRD1 as the main genes responsible for the Williams–Beuren syndrome neurocognitive profile. J Med Genet 47:312–320. doi:10.1136/jmg.2009.071712
Bass-Zubek AE, Godsel LM, Delmar M, Green KJ (2009) Plakophilins: multifunctional scaffolds for adhesion and signaling. Curr Opin Cell Biol 21:708–716. doi:10.1016/j.ceb.2009.07.002
Bayarsaihan D, Ruddle FH (2000) Isolation and characterization of BEN, a member of the TFII-I family of DNA-binding proteins containing distinct helix–loop–helix domains. Proc Natl Acad Sci USA 97:7342–7347
Binder JX, Pletscher-Frankild S, Tsafou K, Stolte C, O’Donoghue SI, Schneider R, Jensen LJ (2014) COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database (Oxford) 2014:bau012. doi:10.1093/database/bau012
Canzio D, Larson A, Narlikar GJ (2014) Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol 24:377–386. doi:10.1016/j.tcb.2014.01.002
Caraveo G, van Rossum DB, Patterson RL, Snyder SH, Desiderio S (2006) Action of TFII-I outside the nucleus as an inhibitor of agonist-induced calcium entry. Science 314:122–125. doi:10.1126/science.1127815
Chen X, Bonne S, Hatzfeld M, van Roy F, Green KJ (2002) Protein binding and functional characterization of plakophilin 2. Evidence for its diverse roles in desmosomes and beta-catenin signaling. J Biol Chem 277:10512–10522. doi:10.1074/jbc.M108765200
Denham M, Dottori M (2011) Neural differentiation of induced pluripotent stem cells. Methods Mol Biol 793:99–110. doi:10.1007/978-1-61779-328-8_7
Denham M, Parish CL, Leaw B, Wright J, Reid CA, Petrou S, Dottori M, Thompson LH (2012) Neurons derived from human embryonic stem cells extend long-distance axonal projections through growth along host white matter tracts after intra-cerebral transplantation. Front Cell Neurosci 6:11. doi:10.3389/fncel.2012.00011
Depienne C, Heron D, Betancur C, Benyahia B, Trouillard O, Bouteiller D, Verloes A, LeGuern E, Leboyer M, Brice A (2007) Autism, language delay and mental retardation in a patient with 7q11 duplication. J Med Genet 44:452–458. doi:10.1136/jmg.2006.047092
Enkhmandakh B, Makeyev AV, Erdenechimeg L, Ruddle FH, Chimge NO, Tussie-Luna MI, Roy AL, Bayarsaihan D (2009) Essential functions of the Williams–Beuren syndrome-associated TFII-I genes in embryonic development. Proc Natl Acad Sci USA 106:181–186. doi:10.1073/pnas.0811531106
Franke Y, Peoples RJ, Francke U (1999) Identification of GTF2IRD1, a putative transcription factor within the Williams–Beuren syndrome deletion at 7q11.23. Cytogenet Genome Res 86:296–304
Fujita N, Watanabe S, Ichimura T, Ohkuma Y, Chiba T, Saya H, Nakao M (2003) MCAF mediates MBD1-dependent transcriptional repression. Mol Cell Biol 23:2834–2843
Gietz RD, Woods RA (2002) Transformation of yeast by LiAc/SS carrier DNA/PEG Method. Methods Enzymol 35:87–96
Golbabapour S, Majid NA, Hassandarvish P, Hajrezaie M, Abdulla MA, Hadi AH (2013) Gene silencing and Polycomb group proteins: an overview of their structure, mechanisms and phylogenetics. OMICS 17:283–296. doi:10.1089/omi.2012.0105
Guemez-Gamboa A, Coufal NG, Gleeson JG (2014) Primary cilia in the developing and mature brain. Neuron 82:511–521. doi:10.1016/j.neuron.2014.04.024
Gunbin KV, Ruvinsky A (2013) Evolution of general transcription factors. J Mol Evol 76:28–47. doi:10.1007/s00239-012-9535-y
Hakimi MA, Dong Y, Lane WS, Speicher DW, Shiekhattar R (2003) A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J Biol Chem 278:7234–7239. doi:10.1074/jbc.M208992200
Han YG, Kim HJ, Dlugosz AA, Ellison DW, Gilbertson RJ, Alvarez-Buylla A (2009) Dual and opposing roles of primary cilia in medulloblastoma development. Nat Med 15:1062–1065. doi:10.1038/nm.2020
Hatzfeld M, Haffner C, Schulze K, Vinzens U (2000) The function of plakophilin 1 in desmosome assembly and actin filament organization. J Cell Biol 149:209–222
Howard ML, Palmer SJ, Taylor KM, Arthurson GJ, Spitzer MW, Du X, Pang TY, Renoir T, Hardeman EC, Hannan AJ (2012) Mutation of Gtf2ird1 from the Williams–Beuren syndrome critical region results in facial dysplasia, motor dysfunction, and altered vocalisations. Neurobiol Dis 45:913–922. doi:10.1016/j.nbd.2011.12.010
Issa LL, Palmer SJ, Guven KL, Santucci N, Hodgson VR, Popovic K, Joya JE, Hardeman EC (2006) MusTRD can regulate postnatal fiber-specific expression. Dev Biol 293:104–115. doi:10.1016/j.ydbio.2006.01.019
Jackson TA, Taylor HE, Sharma D, Desiderio S, Danoff SK (2005) Vascular endothelial growth factor receptor-2: counter-regulation by the transcription factors, TFII-I and TFII-IRD1. J Biol Chem 280:29856–29863. doi:10.1074/jbc.M500335200
Jiang W, Sordella R, Chen GC, Hakre S, Roy AL, Settleman J (2005) An FF domain-dependent protein interaction mediates a signaling pathway for growth factor-induced gene expression. Mol Cell 17:23–35. doi:10.1016/j.molcel.2004.11.024
Lopez-Domenech G, Serrat R, Mirra S, D’Aniello S, Somorjai I, Abad A, Vitureira N, Garcia-Arumi E, Alonso MT, Rodriguez-Prados M, Burgaya F, Andreu AL, Garcia-Sancho J, Trullas R, Garcia-Fernandez J, Soriano E (2012) The Eutherian Armcx genes regulate mitochondrial trafficking in neurons and interact with Miro and Trak2. Nat Commun 3:814. doi:10.1038/ncomms1829
Merla G, Brunetti-Pierri N, Micale L, Fusco C (2010) Copy number variants at Williams–Beuren syndrome 7q11.23 region. Hum Genet 128:3–26. doi:10.1007/s00439-010-0827-2
O’Leary J, Osborne LR (2011) Global analysis of gene expression in the developing brain of Gtf2ird1 knockout mice. PLoS One 6:e23868. doi:10.1371/journal.pone.0023868
O’Mahoney J, Guven KL, Joya JE, Robinson S, Wade RP, Hardeman EC (1998) Identification of a novel slow-muslce-fiber enhancer binding protein, MusTRD1. Mol Cell Biol 18:6641–6652
Osborne LR (2010) Animal models of Williams syndrome. Am J Med Genet C Semin Med Genet 154C:209–219. doi:10.1002/ajmg.c.30257
Osborne LR, Campbell T, Daradich A, Scherer SW, Tsui LC (1999) Identification of a putative transcription factor gene (WBSCR11) that is commonly deleted in Williams–Beuren syndrome. Genomics 57:279–284
Palmer SJ, Tay ES, Santucci N, Cuc Bach TT, Hook J, Lemckert FA, Jamieson RV, Gunnning PW, Hardeman EC (2007) Expression of Gtf2ird1, the Williams syndrome-associated gene, during mouse development. Gene Expr Patterns 7:396–404. doi:10.1016/j.modgep.2006.11.008
Palmer SJ, Santucci N, Widagdo J, Bontempo SJ, Taylor KM, Tay ES, Hook J, Lemckert F, Gunning PW, Hardeman EC (2010) Negative autoregulation of GTF2IRD1 in Williams–Beuren syndrome via a novel DNA binding mechanism. J Biol Chem 285:4715–4724. doi:10.1074/jbc.M109.086660
Palmer SJ, Taylor KM, Santucci N, Widagdo J, Chan YK, Yeo JL, Adams M, Gunning PW, Hardeman EC (2012) GTF2IRD2 from the Williams–Beuren critical region encodes a mobile-element-derived fusion protein that antagonizes the action of its related family members. J Cell Sci 125:5040–5050. doi:10.1242/jcs.102798
Pérez Jurado LA, Wang Y-K, Peoples R, Coloma A, Cruces J, Francke U (1998) A duplicated gene in the breakpoint regions of the 7q11.23 Williams-Beuren syndrome deletion encodes the initiator binding protein TFII-I and BAP-135, a phosphorylation target of BTK. Hum Mol Genet 7:325–334
Polly P, Haddadi LM, Issa LL, Subramaniam N, Palmer SJ, Tay ES, Hardeman EC (2003) hMusTRD1a1 represses MEF2 activation of the troponin I slow enhancer. J Biol Chem 278:36603–36610
Proulx E, Young EJ, Osborne LR, Lambe EK (2010) Enhanced prefrontal serotonin 5-HT(1A) currents in a mouse model of Williams–Beuren syndrome with low innate anxiety. J Neurodev Disord 2:99–108. doi:10.1007/s11689-010-9044-5
Ring C, Ogata S, Meek L, Song J, Ohta T, Miyazono K, Cho KW (2002) The role of a Williams–Beuren syndrome-associated helix–loop–helix domain-containing transcription factor in activin/nodal signaling. Genes Dev 16:820–835. doi:10.1101/gad.963802
Roy AL (2012) Biochemistry and biology of the inducible multifunctional transcription factor TFII-I: 10 years later. Gene 492:32–41. doi:10.1016/j.gene.2011.10.030
Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, Mason CE, Bilguvar K, Celestino-Soper PB, Choi M, Crawford EL, Davis L, Wright NR, Dhodapkar RM, DiCola M, DiLullo NM, Fernandez TV, Fielding-Singh V, Fishman DO, Frahm S, Garagaloyan R, Goh GS, Kammela S, Klei L, Lowe JK, Lund SC, McGrew AD, Meyer KA, Moffat WJ, Murdoch JD, O’Roak BJ, Ober GT, Pottenger RS, Raubeson MJ, Song Y, Wang Q, Yaspan BL, Yu TW, Yurkiewicz IR, Beaudet AL, Cantor RM, Curland M, Grice DE, Gunel M, Lifton RP, Mane SM, Martin DM, Shaw CA, Sheldon M, Tischfield JA, Walsh CA, Morrow EM, Ledbetter DH, Fombonne E, Lord C, Martin CL, Brooks AI, Sutcliffe JS, Cook EH Jr, Geschwind D, Roeder K, Devlin B, State MW (2011) Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 70:863–885. doi:10.1016/j.neuron.2011.05.002
Schneider T, Skitt Z, Liu Y, Deacon RM, Flint J, Karmiloff-Smith A, Rawlins JN, Tassabehji M (2012) Anxious, hypoactive phenotype combined with motor deficits in Gtf2ird1 null mouse model relevant to Williams syndrome. Behav Brain Res 233:458–473. doi:10.1016/j.bbr.2012.05.014
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi:10.1101/gr.1239303
Sleeman JE, Trinkle-Mulcahy L (2014) Nuclear bodies: new insights into assembly/dynamics and disease relevance. Curr Opin Cell Biol 28:76–83. doi:10.1016/j.ceb.2014.03.004
Somerville MJ, Mervis CB, Young EJ, Seo EJ, del Campo M, Bamforth S, Peregrine E, Loo W, Lilley M, Perez-Jurado LA, Morris CA, Scherer SW, Osborne LR (2005) Severe expressive-language delay related to duplication of the Williams–Beuren locus. N Engl J Med 353:1694–1701. doi:10.1056/NEJMoa051962
Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M, Carey M, Garcia BA, Plath K (2013) Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1gamma in reprogramming to pluripotency. Nat Cell Biol 15:872–882. doi:10.1038/ncb2768
Tanikawa M, Wada-Hiraike O, Nakagawa S, Shirane A, Hiraike H, Koyama S, Miyamoto Y, Sone K, Tsuruga T, Nagasaka K, Matsumoto Y, Ikeda Y, Shoji K, Oda K, Fukuhara H, Nakagawa K, Kato S, Yano T, Taketani Y (2011) Multifunctional transcription factor TFII-I is an activator of BRCA1 function. Br J Cancer 104:1349–1355. doi:10.1038/bjc.2011.75
Tantin D, Tussie-Luna MI, Roy AL, Sharp PA (2004) Regulation of immunoglobulin promoter activity by TFII-I class transcription factors. J Biol Chem 279:5460–5469. doi:10.1074/jbc.M311177200
Tassabehji M, Carette M, Wilmot C, Donnai D, Read AP, Metcalfe K (1999) A transcription factor involved in skeletal muscle gene expression is deleted in patients with Williams syndrome. Eur J Hum Genet 7:737–747. doi:10.1038/sj.ejhg.5200396
Tassabehji M, Hammond P, Karmiloff-Smith A, Thompson P, Thorgeirsson SS, Durkin ME, Popescu NC, Hutton T, Metcalfe K, Rucka A, Stewart H, Read AP, Maconochie M, Donnai D (2005) GTF2IRD1 in craniofacial development of humans and mice. Science 310:1184–1187. doi:10.1126/science.1116142
Tay ES, Guven KL, Subramaniam N, Polly P, Issa LL, Gunning PW, Hardeman EC (2003) Regulation of alternative splicing of Gtf2ird1 and its impact on slow muscle promoter activity. Biochem J 374:359–367. doi:10.1042/BJ20030189
Thompson PD, Webb M, Beckett W, Hinsley T, Jowitt T, Sharrocks AD, Tassabehji M (2007) GTF2IRD1 regulates transcription by binding an evolutionarily conserved DNA motif ‘GUCE’. FEBS Lett 581:1233–1242. doi:10.1016/j.febslet.2007.02.040
Tipney HJ, Hinsley TA, Brass A, Metcalfe K, Donnai D, Tassabehji M (2004) Isolation and characterisation of GTF2IRD2, a novel fusion gene mapping to the Williams–Beuren syndrome critical region. Eur J Hum Genet 12:551–560
Torniero C, dalla Bernardina B, Novara F, Vetro A, Ricca I, Darra F, Pramparo T, Guerrini R, Zuffardi O (2007) Cortical dysplasia of the left temporal lobe might explain severe expressive-language delay in patients with duplication of the Williams–Beuren locus. Eur J Hum Genet 15:62–67. doi:10.1038/sj.ejhg.5201730
Tussie-Luna MI, Michel B, Hakre S, Roy AL (2002) The SUMO ubiquitin-protein isopeptide ligase family member Miz1/PIASxbeta/Siz2 is a transcriptional cofactor for TFII-I. J Biol Chem 277:43185–43193. doi:10.1074/jbc.M207635200
Van der Aa N, Rooms L, Vandeweyer G, van den Ende J, Reyniers E, Fichera M, Romano C, Delle Chiaie B, Mortier G, Menten B, Destree A, Maystadt I, Mannik K, Kurg A, Reimand T, McMullan D, Oley C, Brueton L, Bongers EM, van Bon BW, Pfund R, Jacquemont S, Ferrarini A, Martinet D, Schrander-Stumpel C, Stegmann AP, Frints SG, de Vries BB, Ceulemans B, Kooy RF (2009) Fourteen new cases contribute to the characterization of the 7q11.23 microduplication syndrome. Eur J Med Genet 52:94–100. doi:10.1016/j.ejmg.2009.02.006
Vullhorst D, Buonanno A (2003) Characterisation of general transcription factor 3, a transcription factor involved in slow muscle-specific gene expression. J Biol Chem 278:8370–8379. doi:10.1074/jbc.M209361200
Vullhorst D, Buonanno A (2005) Multiple GTF2I-like repeats of general transcription factor 3 exhibit DNA binding properties. Evidence for a common origin as a sequence-specific DNA interaction module. J Biol Chem 280:31722–31731. doi:10.1074/jbc.M500593200
Widagdo J, Taylor KM, Gunning PW, Hardeman EC, Palmer SJ (2012) SUMOylation of GTF2IRD1 regulates protein partner interactions and ubiquitin-mediated degradation. PLoS One 7:e49283. doi:10.1371/journal.pone.0049283
Yang W, Desiderio S (1997) BAP-135, a target for Bruton’s tyrosine kinase in response to B cell receptor engagement. Proc Natl Acad Sci USA 94:604–609
Young EJ, Lipina T, Tam E, Mandel A, Clapcote SJ, Bechard AR, Chambers J, Mount HT, Fletcher PJ, Roder JC, Osborne LR (2008) Reduced fear and aggression and altered serotonin metabolism in Gtf2ird1-targeted mice. Genes Brain Behav 7:224–234. doi:10.1111/j.1601-183X.2007.00343.x
Acknowledgments
We thank Kylie M. Taylor for her technical assistance. We are grateful for the plasmid constructs provided by the researchers detailed in the Supplementary Table 1. We extend thanks to the Biomedical Imaging Facility, from the Mark Wainwright Analytical Centre at UNSW Australia for their training and support for the microscopy techniques. PC-M and CPC are recipients of a CONICYT-Becas Chile scholarship from the Government of Chile.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was supported by the National Health and Medical Research Council of Australia (Project Grant 1049639).
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Carmona-Mora, P., Widagdo, J., Tomasetig, F. et al. The nuclear localization pattern and interaction partners of GTF2IRD1 demonstrate a role in chromatin regulation. Hum Genet 134, 1099–1115 (2015). https://doi.org/10.1007/s00439-015-1591-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-015-1591-0