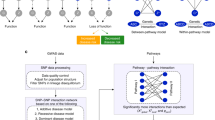
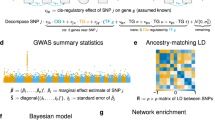
Abstract
Millions of genetic variants have been assessed for their effects on the trait of interest in genome-wide association studies (GWAS). The complex traits are affected by a set of inter-related genes. However, the typical GWAS only examine the association of a single genetic variant at a time. The individual effects of a complex trait are usually small, and the simple sum of these individual effects may not reflect the holistic effect of the genetic system. High-throughput methods enable genomic studies to produce a large amount of data to expand the knowledge base of the biological systems. Biological networks and pathways are built to represent the functional or physical connectivity among genes. Integrated with GWAS data, the network- and pathway-based methods complement the approach of single genetic variant analysis, and may improve the power to identify trait-associated genes. Taking advantage of the biological knowledge, these approaches are valuable to interpret the functional role of the genetic variants, and to further understand the molecular mechanism influencing the traits. The network- and pathway-based methods have demonstrated their utilities, and will be increasingly important to address a number of challenges facing the mainstream GWAS.


Similar content being viewed by others
References
Abatangelo L, Maglietta R, Distaso A, D’Addabbo A, Creanza T, Mukherjee S, Ancona N (2009) Comparative study of gene set enrichment methods. BMC Bioinformatics 10:275
Akula N, Baranova A, Seto D, Solka J, Nalls MA, Singleton A, Ferrucci L, Tanaka T, Bandinelli S, Cho YS, Kim YJ, Lee JY, Han BG, Bipolar Disorder Genome Study Cconsortium; Wellcome Trust Case-Control Cconsortium, McMahon FJ (2011) A network-based approach to prioritize results from genome-wide association studies. PLoS One 6:e24220. doi:10.1371/journal.pone.0024220
Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, Kerssemakers J, Leroy C, Menden M, Michaut M, Montecchi-Palazzi L, Neuhauser SN, Orchard S, Perreau V, Roechert B, van Eijk K, Hermjakob H (2010) The IntAct molecular interaction database in 2010. Nucleic Acids Res 38:D525–D531. doi:10.1093/nar/gkp878
Askland K, Read C, Moore J (2009) Pathways-based analyses of whole-genome association study data in bipolar disorder reveal genes mediating ion channel activity and synaptic neurotransmission. Hum Genet 125:63–79. doi:10.1007/s00439-008-0600-y
Askland K, Read C, O’Connell C, Moore JH (2012) Ion channels and schizophrenia: a gene set-based analytic approach to GWAS data for biological hypothesis testing. Hum Genet 131:373–391. doi:10.1007/s00439-011-1082-x
Bansal V, Libiger O, Torkamani A, Schork NJ (2010) Statistical analysis strategies for association studies involving rare variants. Nat Rev Genet 11:773–785. doi:10.1038/nrg2867
Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Holko M, Ayanbule O, Yefanov A, Soboleva A (2011) NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res 39:D1005–D1010. doi:10.1093/nar/gkq1184
Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Paley S, Popescu L, Pujar A, Shearer AG, Zhang P, Karp PD (2010) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 38:D473–D479. doi:10.1093/nar/gkp875
Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE (2008) Variations in DNA elucidate molecular networks that cause disease. Nature 452:429–435. doi:10.1038/nature06757
Chen LS, Hutter CM, Potter JD, Liu Y, Prentice RL, Peters U, Hsu L (2010) Insights into colon cancer etiology via a regularized approach to gene set analysis of GWAS data. Am J Hum Genet 86:860–871. doi:10.1016/j.ajhg.2010.04.014
Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD (2007) Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2:2366–2382. doi:10.1038/nprot.2007.324
Davis NA, Crowe JE Jr, Pajewski NM, McKinney BA (2010) Surfing a genetic association interaction network to identify modulators of antibody response to smallpox vaccine. Genes Immun 11:630–636. doi:10.1038/gene.2010.37
Dering C, Hemmelmann C, Pugh E, Ziegler A (2011) Statistical analysis of rare sequence variants: an overview of collapsing methods. Genet Epidemiol 35(Suppl 1):S12–S17. doi:10.1002/gepi.20643
Dobrin R, Zhu J, Molony C, Argman C, Parrish ML, Carlson S, Allan MF, Pomp D, Schadt EE (2009) Multi-tissue coexpression networks reveal unexpected subnetworks associated with disease. Genome Biol 10:R55. doi:10.1186/gb-2009-10-5-r55
Efron B, Tibshirani R (2007) On testing the significance of sets of genes. Ann Appl Stat 1:107–129
Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, Mouy M, Steinthorsdottir V, Eiriksdottir GH, Bjornsdottir G, Reynisdottir I, Gudbjartsson D, Helgadottir A, Jonasdottir A, Jonasdottir A, Styrkarsdottir U, Gretarsdottir S, Magnusson KP, Stefansson H, Fossdal R, Kristjansson K, Gislason HG, Stefansson T, Leifsson BG, Thorsteinsdottir U, Lamb JR, Gulcher JR, Reitman ML, Kong A, Schadt EE, Stefansson K (2008) Genetics of gene expression and its effect on disease. Nature 452:423–428. doi:10.1038/nature06758
Gamazon ER, Zhang W, Konkashbaev A, Duan S, Kistner EO, Nicolae DL, Dolan ME, Cox NJ (2010) SCAN: SNP and copy number annotation. Bioinformatics 26:259–262. doi:10.1093/bioinformatics/btp644
Gandhi TK, Zhong J, Mathivanan S, Karthick L, Chandrika KN, Mohan SS, Sharma S, Pinkert S, Nagaraju S, Periaswamy B, Mishra G, Nandakumar K, Shen B, Deshpande N, Nayak R, Sarker M, Boeke JD, Parmigiani G, Schultz J, Bader JS, Pandey A (2006) Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat Genet 38:285–293
Ghazalpour A, Doss S, Zhang B, Wang S, Plaisier C, Castellanos R, Brozell A, Schadt EE, Drake TA, Lusis AJ, Horvath S (2006) Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet 2:e130. doi:10.1371/journal.pgen.0020130
Goeman JJ, Buhlmann P (2007) Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics 23:980–987. doi:10.1093/bioinformatics/btm051
Hannum G, Srivas R, Guenole A, van Attikum H, Krogan NJ, Karp RM, Ideker T (2009) Genome-wide association data reveal a global map of genetic interactions among protein complexes. PLoS Genet 5:e1000782. doi:10.1371/journal.pgen.1000782
Holden M, Deng S, Wojnowski L, Kulle B (2008) GSEA-SNP: applying gene set enrichment analysis to SNP data from genome-wide association studies. Bioinformatics 24:2784
Holmans P (2010) Statistical methods for pathway analysis of genome-wide data for association with complex genetic traits. Adv Genet 72:141–179. doi:10.1016/B978-0-12-380862-2.00007-2
Holmans P, Green EK, Pahwa JS, Ferreira MA, Purcell SM, Sklar P, Wellcome Trust Case–Control C, Owen MJ, O’Donovan MC, Craddock N (2009) Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am J Hum Genet 85: 13–24. doi:10.1016/j.ajhg.2009.05.011
Hu T, Sinnott-Armstrong NA, Kiralis JW, Andrew AS, Karagas MR, Moore JH (2011) Characterizing genetic interactions in human disease association studies using statistical epistasis networks. BMC Bioinformatics 12:364. doi:10.1186/1471-2105-12-364
Ideker T, Krogan NJ (2012) Differential network biology. Mol Syst Biol 8:565. doi:10.1038/msb.2011.99
Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292:929
International Consortium for Blood Pressure Genome-Wide Association Studies, Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O’Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sober S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der Harst P, Kao WH, Sjogren M, Vinay DG, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimaki T, Matullo G, Wu Y, Gaunt TR, Onland-Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND, CARDIoGRAM consortium; CKDGen Consortium; KidneyGen Consortium; EchoGen consortium; CHARGE-HF consortium,, Aspelund T, Garcia M, Chang YP, O’Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, et al (2011) Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478: 103-9. doi:10.1038/nature10405
Jia P, Zheng S, Long J, Zheng W, Zhao Z (2011) dmGWAS: dense module searching for genome-wide association studies in protein–protein interaction networks. Bioinformatics 27:95–102. doi:10.1093/bioinformatics/btq615
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M, Kashyap MK, Mohmood R, Ramachandra YL, Krishna V, Rahiman BA, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A (2009) Human protein reference database–2009 update. Nucleic Acids Res 37:D767–D772. doi:10.1093/nar/gkn892
Kraft P, Raychaudhuri S (2009) Complex diseases, complex genes: keeping pathways on the right track. Epidemiology 20:508–511. doi:10.1097/EDE.0b013e3181a93b98
Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM (2011) Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res 21:1109–1121. doi:10.1101/gr.118992.110
Luo L, Peng G, Zhu Y, Dong H, Amos CI, Xiong M (2010) Genome-wide gene and pathway analysis. Eur J Hum Genet 18:1045–1053. doi:http://www.nature.com/ejhg/journal/v18/n9/suppinfo/ejhg201062s1.html
Ma L, Brautbar A, Boerwinkle E, Sing CF, Clark AG, Keinan A (2012) Knowledge-driven analysis identifies a gene–gene interaction affecting high-density lipoprotein cholesterol levels in multi-ethnic populations. PLoS Genet 8:e1002714. doi:10.1371/journal.pgen.1002714
Moore JH, Williams SM (2009) Epistasis and its implications for personal genetics. Am J Hum Genet 85:309–320
Newman MEJ (2010) Networks: an introduction. Oxford University Press, Oxford, New York
Newton MA, Quintana FA, den Boon JA, Sengupta S, Ahlquist P (2007) Random-set methods identify distinct aspects of the enrichment signal in gene-set analysis. Ann Appl Stat 1:85–106
Pagel P, Kovac S, Oesterheld M, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Mark P, Stumpflen V, Mewes HW, Ruepp A, Frishman D (2005) The MIPS mammalian protein–protein interaction database. Bioinformatics (Oxford, England) 21:832–834
Pattin KA, Moore JH (2008) Exploiting the proteome to improve the genome-wide genetic analysis of epistasis in common human diseases. Hum Genet 124:19–29
Pattin KA, Moore JH (2009) Role for protein–protein interaction databases in human genetics. Expert Rev Proteomics 6:647–659
Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, Carninci P, Daub CO, Forrest AR, Gough J, Grimmond S, Han JH, Hashimoto T, Hide W, Hofmann O, Kamburov A, Kaur M, Kawaji H, Kubosaki A, Lassmann T, van Nimwegen E, MacPherson CR, Ogawa C, Radovanovic A, Schwartz A, Teasdale RD, Tegner J, Lenhard B, Teichmann SA, Arakawa T, Ninomiya N, Murakami K, Tagami M, Fukuda S, Imamura K, Kai C, Ishihara R, Kitazume Y, Kawai J, Hume DA, Ideker T, Hayashizaki Y (2010) An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140:744–752. doi:10.1016/j.cell.2010.01.044
Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, International Inflammatory Bowel Disease Genetics Consorium, Cotsapas C, Daly MJ (2011) Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet 7:e1001273. doi:10.1371/journal.pgen.1001273
Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, Zhu J, Millstein J, Sieberts S, Lamb J, GuhaThakurta D, Derry J, Storey JD, Avila-Campillo I, Kruger MJ, Johnson JM, Rohl CA, van Nas A, Mehrabian M, Drake TA, Lusis AJ, Smith RC, Guengerich FP, Strom SC, Schuetz E, Rushmore TH, Ulrich R (2008) Mapping the genetic architecture of gene expression in human liver. PLoS Biol 6:e107. doi:10.1371/journal.pbio.0060107
Schaid DJ, Sinnwell JP, Jenkins GD, McDonnell SK, Ingle JN, Kubo M, Goss PE, Costantino JP, Wickerham DL, Weinshilboum RM (2012) Using the gene ontology to scan multilevel gene sets for associations in genome wide association studies. Genet Epidemiol 36:3–16. doi:10.1002/gepi.20632
Segre AV, Consortium D, investigators M, Groop L, Mootha VK, Daly MJ, Altshuler D (2010) Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet 6. doi:10.1371/journal.pgen.1001058
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi:10.1101/gr.1239303
Sieberts SK, Schadt EE (2007) Moving toward a system genetics view of disease. Mamm Genome 18:389–401. doi:10.1007/s00335-007-9040-6
Sowa ME, Bennett EJ, Gygi SP, Harper JW (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403
Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34:D535–D539
Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE (2005) A human protein–protein interaction network: a resource for annotating the proteome. Cell 122:957–968
Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, Lyle R, Hunt S, Kahl B, Antonarakis SE, Tavare S, Deloukas P, Dermitzakis ET (2005) Genome-wide associations of gene expression variation in humans. PLoS Genet 1:e78
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545–15550. doi:10.1073/pnas.0506580102
Sun YV, Kardia SL (2010) Identification of epistatic effects using a protein–protein interaction database. Hum Mol Genet 19:4345–4352. doi:10.1093/hmg/ddq356
Sun YV, Sung YJ, Tintle N, Ziegler A (2011) Identification of genetic association of multiple rare variants using collapsing methods. Genet Epidemiol 35(Suppl 1):S101–S106. doi:10.1002/gepi.20658
Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM, Broad GO, Seattle GO, on behalf of the NESP (2012) Evolution and Functional Impact of Rare Coding Variation from Deep Sequencing of Human Exomes. Science. doi:10.1126/science.1219240
Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI et al (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466:707–713. doi:10.1038/nature09270
Thomas DC (2000) Genetic epidemiology with a capital“E”. Genet Epidemiol 19:289–300
Thomas DC (2012) Genetic epidemiology with a capital E: where will we be in another 10 years? Genetic Epidemiology. doi:10.1002/gepi.21612
Wang K, Li M, Bucan M (2007) Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet 81:1278–1283. doi:10.1086/522374
Wang K, Li M, Hakonarson H (2010) Analysing biological pathways in genome-wide association studies. Nat Rev Genet 11:843–854. doi:10.1038/nrg2884
Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38:W214–W220. doi:10.1093/nar/gkq537
Weng L, Macciardi F, Subramanian A, Guffanti G, Potkin SG, Yu Z, Xie X (2011) SNP-based pathway enrichment analysis for genome-wide association studies. BMC Bioinformatics 12:99. doi:10.1186/1471-2105-12-99
Xenarios I, Salwinski L, Duan XJ, Higney P, Kim SM, Eisenberg D (2002) DIP, the database of interacting proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res 30:303–305
Yu H, Tardivo L, Tam S, Weiner E, Gebreab F, Fan C, Svrzikapa N, Hirozane-Kishikawa T, Rietman E, Yang X, Sahalie J, Salehi-Ashtiani K, Hao T, Cusick ME, Hill DE, Roth FP, Braun P, Vidal M (2011) Next-generation sequencing to generate interactome datasets. Nat Methods 8:478–480. doi:10.1038/nmeth.1597
Zhang K, Cui S, Chang S, Zhang L, Wang J (2010) i-GSEA4GWAS: a web server for identification of pathways/gene sets associated with traits by applying an improved gene set enrichment analysis to genome-wide association study. Nucleic Acids Res 38:W90–W95. doi:10.1093/nar/gkq324
Zhong H, Beaulaurier J, Lum PY, Molony C, Yang X, Macneil DJ, Weingarth DT, Zhang B, Greenawalt D, Dobrin R, Hao K, Woo S, Fabre-Suver C, Qian S, Tota MR, Keller MP, Kendziorski CM, Yandell BS, Castro V, Attie AD, Kaplan LM, Schadt EE (2010) Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet 6:e1000932. doi:10.1371/journal.pgen.1000932
Zhu J, Zhang B, Smith EN, Drees B, Brem RB, Kruglyak L, Bumgarner RE, Schadt EE (2008) Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat Genet 40:854–861. doi:10.1038/ng.167
Zuk O, Hechter E, Sunyaev SR, Lander ES (2012) The mystery of missing heritability: genetic interactions create phantom heritability. Proc Natl Acad Sci USA. doi:10.1073/pnas.1119675109
Acknowledgments
This study was partly supported by the National Institute of Health grant HL100245.
Conflict of interest
The author declares that he has no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sun, Y.V. Integration of biological networks and pathways with genetic association studies. Hum Genet 131, 1677–1686 (2012). https://doi.org/10.1007/s00439-012-1198-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-012-1198-7