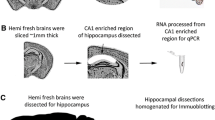
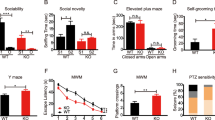
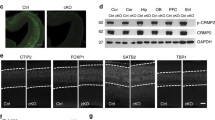
Abstract
Coffin–Lowry syndrome (CLS) is a syndromic form of mental retardation caused by loss of function mutations in the X-linked RPS6KA3 gene, which encodes RSK2, a serine/threonine kinase acting in the MAPK/ERK pathway. The mouse invalidated for the Rps6ka3 (Rsk2-KO) gene displays learning and long-term spatial memory deficits. In the current study, we compared hippocampal gene expression profiles from Rsk2-KO and normal littermate mice to identify changes in molecular pathways. Differential expression was observed for 100 genes encoding proteins acting in various biological pathways, including cell growth and proliferation, cell death and higher brain function. The twofold up-regulated gene (Gria2) was of particular interest because it encodes the subunit GLUR2 of the AMPA glutamate receptor. AMPA receptors mediate most fast excitatory synaptic transmission in the central nervous system. We provide evidence that in the hippocampus of Rsk2-KO mice, expression of GLUR2 at the mRNA and at the protein levels is significantly increased, whereas basal AMPA receptor-mediated transmission in the hippocampus of Rsk2-KO mice is significantly decreased. This is the first time that such deregulations have been demonstrated in the mouse model of the Coffin–Lowry syndrome. Our findings suggest that a defect in AMPA neurotransmission and plasticity contribute to mental retardation in CLS patients.






Similar content being viewed by others
References
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300
Birnbaumer L, Qin N, Olcese R, Tareilus E, Platano D, Costantin J, Stefani E (1998) Structures and functions of calcium channel beta subunits. J Bioenerg Biomembr 30:357–375
Borges K, Dingledine R (2001) Functional organization of the GluR1 glutamate receptor promoter. J Biol Chem 276:25929–25938
Brand K, Page S, Walli AK, Neumeier D, Baeuerle PA (1997) Role of nuclear factor-kappa B in atherogenesis. Exp Physiol 82:297–304
Brorson JR, Li D, Suzuki T (2004) Selective expression of heteromeric AMPA receptors driven by flip-flop differences. J Neurosci 24:3461–3470
Carlson NG, Howard J, Gahring LC, Rogers SW (2000) RNA editing (Q/R site) and flop/flip splicing of AMPA receptor transcripts in young and old brains. Neurobiol Aging 21:599–606
Cheng CM, Mervis RF, Niu SL, Salem N Jr, Witters LA, Tseng V, Reinhardt R, Bondy CA (2003) Insulin-like growth factor 1 is essential for normal dendritic growth. J Neurosci Res 73:1–9
Davis S, Laroche S (2006) Mitogen-activated protein kinase/extracellular regulated kinase signalling and memory stabilization: a review. Genes Brain Behav 5(Suppl 2):61–72
De Cesare D, Jacquot S, Hanauer A, Sassone-Corsi P (1998) Rsk-2 activity is necessary for epidermal growth factor-induced phosphorylation of CREB protein and transcription of c-fos gene. Proc Nat Acad Sci USA 95:12202–12207
Frödin M, Gammeltoft S (1999) Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol Cell Endocrinol 51:65–77
Fu AK, Cheung WM (1999) Identification of genes induced by neuregulin in cultured myotubes. Mol Cell Neurosci 14:241–253
Ghate A, Befort K, Becker JA, Filliol D, Bole-Feysot C, Demebele D, Jost B, Koch M, Kieffer BL (2007) Identification of novel striatal genes by expression profiling in adult mouse brain. Neuroscience 146:1182–1192
Guimiot F, Delezoide AL, Hanauer A, Simonneau M (2004) Expression of the RSK2 gene during early human development. Gene Expr Patterns 4:111–114
Hanauer A, Young ID (2002) Coffin–Lowry syndrome: clinical and molecular features. J Med Genet 39:705–713
Isaac JT, Ashby M, McBain CJ (2007) The role of the GluR2 subunit in AMPA receptor function and synaptic plasticity. Neuron 54:859–871
Kessels HW, Malinow R (2009) Synaptic AMPA receptor plasticity and behavior. Neuron 61:340–350
Kim MD, Jan LY, Jan YN (2006) The bHLH-PAS protein Spineless is necessary for the diversification of dendrite morphology of Drosophila dendritic arborization neurons. Genes Dev 20:2773–2778
Kuner T, Beck C, Sakmann B, Seeburg PH (2001) Channel-lining residues of the AMPA receptor M2 segment: structural environment of the Q/R site and identification of the selectivity filter. J Neurosci 21:4162–4172
Lai F, Chen CX, Carter KC, Nishikura K (1997) Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol Cell Biol 17:2413–2424
Lee JC, Greig A, Ravindranathan A, Parks TN, Rao MS (1998) Molecular analysis of AMPA-specific receptors: subunit composition, editing, and calcium influx determination in small amounts of tissue. Brain Res Protoc 3:142–154
Lomeli H, Mosbacher J, Melcher T, Höger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH (1994) Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266:1709–1713
Lutz RE (2007) Trinucleotide repeat disorders. Semin Pediatr Neurol 14:26–33
Malinow R, Malenka RC (2002) AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci 25:103–126
Marques Pereira P, Gruss M, Braun K, Foos N, Pannetier S, Hanauer A (2008) Dopaminergic system dysregulation in the mrsk2_KO mouse, an animal model of the Coffin–Lowry syndrome. J Neurochem 107:1325–1334
Nakamoto N, Nalavadi V, Epstein MP, Narayanan U, Bassell GJ, Warren ST (2007) Fragile X mental retardation protein deficiency leads to excessive mGluR5-dependent internalization of AMPA receptors. Proc Nat Acad Sci USA 39:15537–15542
Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, Mora M, Riggs JE, Oh SJ, Koga Y et al (2000) Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 406:906–910
Passafaro M, Piëch V, Sheng M (2001) Subunit-specific temporal and spatial patterns of AMPA receptor exocytosis in hippocampal neurons. Nat Neurosci 4:917–926
Pei W, Huang Z, Wang C, Han Y, Park JS, Niu L (2009) Flip and flop: a molecular determinant for AMPA receptor channel opening. Biochemistry 48:3767–3777
Peng PL, Zhong X, Tu W, Soundarapandian MM, Molner P, Zhu D, Lau L, Liu S, Liu F, Lu Y (2006) ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron 49:719–733
Poirier R, Jacquot S, Vaillend C, Soutthiphong AA, Libbey M, Davis S, Laroche S, Hanauer A, Welzl H, Lipp HP, Wolfer DP (2007) Deletion of the Coffin–Lowry syndrome gene Rsk2 in mice is associated with impaired spatial learning and reduced control of exploratory behavior. Behav Genet 37:31–50
Qin H, Powell-Coffman JA (2004) The Caenorhabditis elegans aryl hydrocarbon receptor, AHR-1, regulates neuronal development. Dev Biol 270:64–75
Rouach N, Byrd K, Petralia RS, Elias GM, Adesnik H, Tomita S, Karimzadegan S, Kealey C, Bredt DS, Nicoll RA (2005) TARP gamma-8 controls hippocampal AMPA receptor number, distribution and synaptic plasticity. Nat Neurosci 8:1525–1533
Sassone-Corsi P, Mizzen CA, Cheung P, Crosio C, Monaco L, Jacquot S, Hanauer A, Allis CD (1999) Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science 285:886–891
Seidenman KJ, Steinberg JP, Huganir R, Malinow R (2003) Glutamate receptor subunit 2 serine 880 phosphorylation modulates synaptic transmission and mediates plasticity in CA1 pyramidal cells. J Neurosci 23:9220–9228
Thomas GM, Rumbaugh GR, Harrar DB, Huganir RL (2005) Ribosomal S6 kinase 2 interacts with and phosphorylates PDZ domain-containing proteins and regulates AMPA receptor transmission. Proc Nat Acad Sci USA 102:15006–15011
Wang Y, Tang BL (2006) SNAREs in neurons—beyond synaptic vesicle exocytosis. Mol Membr Biol 23:377–384
Xu D, Hopf C, Reddy R, Cho RW, Guo L, Lanahan A, Petralia RS, Wenthold RJ, O’Brien RJ, Worley P (2003) Narp and NP1 form heterocomplexes that function in developmental and activity-dependent synaptic plasticity. Neuron 39:513–528
Zeniou M, Ding T, Trivier E, Hanauer A (2002) Expression analysis of RSK gene family members: the RSK2 gene, mutated in Coffin–Lowry syndrome, is prominently expressed in brain structures essential for cognitive function and learning. Hum Mol Genet 11:2929–2940
Zou Y, Liu Q, Chen B, Zhang X, Guo C, Zhou H, Li J, Gao G, Guo Y, Yan C et al (2007) Mutation in CUL4B, which encodes a member of cullin-RING ubiquitin ligase complex, causes X-linked mental retardation. Am J Hum Genet 80:561–566
Acknowledgments
The authors wish to thank Jessica Heringer for technical assistance and helpful discussions, and the IGBMC, Institut de Génétique et de Biologie Moléculaire et Cellulaire for assistance. This work was supported by the French National Agency for Research (Grant number 08-MNPS-021-01, to A.H.), the Fondation Jérôme Lejeune (to A.H.), the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche, the College de France and the University of Strasbourg. AS was supported by a fellowship from the Ministere pour la Recherche et Technologie of France and TM by a scholarship from the Higher Education Commission (HEC) of Pakistan.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
T. Mehmood and A. Schneider contributed equally to this work.
An erratum to this article can be found at http://dx.doi.org/10.1007/s00439-010-0932-2
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1(a) WT and Rsk2-KO hippocampal neurons were labeled with N-terminal GLUR1 antibody under non-permeabilized condition to stain surface GLUR1 (sGLUR1), followed by PSD95 staining (a post-synaptic marker). Arrows point to post-synaptic surface-expressed GLUR1. (b) No significant difference for GLUR1 surface expression was found between WT and Rsk2-KO mice (n = 4 WT and 6 KO embryos).
Supplemental Fig. 2 Determination of relative Gria2 R/G editing and Flip/Flop splice levels. The Gria1 (encoding GLUR1) and Gria2 (encoding GLUR2) mRNAs from five Rsk2-KO and five WT hippocampi were amplified by RT-PCR in triplicates and the products sequenced. Representative chromatograms from one KO and one WT littermate is shown. (a)Gria1 for WT, (b)Gria1 for KO, (c)Gria2 for WT, (d)Gria2 for KO. * points the G/R editing site (A: unedited, G: edited). The peak intensity of the G nucleotide signal was measured and reported as the mean percentage for the five mice of the total signal (A + G). ** points the first nucleotide of the flip/flop exon (flop starts with A and flip with C). The peak intensity of the first nucleotide between the two variants was measured and reported as the mean percentage for the five mice of the total signal (A + C).
Supplemental Fig. 1
(a) WT and Rsk2-KO hippocampal neurons were labeled with N-terminal GLUR1 antibody under non permeabilized condition to stain surface GLUR1 (sGLUR1), followed by PSD95 staining (a post-synaptic marker). Arrows point to post-synaptic surface expressed GLUR1. (b) No significant difference for GLUR1 surface expression was found between WT and Rsk2-KO mice (n= 4 WT and 6 KO embryos) (jpg 303 kb)
Supplemental Fig. 2
Determination of relative Gria2 R/G editing and Flip/Flop splice levels. The Gria1 (encoding GLUR1) and Gria2 (encoding GLUR2) mRNAs from five Rsk2-KO and five WT hippocampi were amplified by RT-PCR in triplicates and the products sequenced. Representative chromatograms from one KO and one WT littermate is shown. (a) Gria1 for WT, (b) Gria1 for KO, (c) Gria2 for WT, (d) Gria2 for KO. * points the G/R editing site (A: unedited, G: edited). The peak intensity of the G nucleotide signal was measured and reported as the mean percentage for the five mice of the total signal (A + G). ** points the first nucleotide of the flip/flop exon (flop starts with A and flip with C). The peak intensity of the first nucleotide between the two variants was measured and reported as the mean percentage for the five mice of the total signal (A + C) (jpg 1090 kb)
Rights and permissions
About this article
Cite this article
Mehmood, T., Schneider, A., Sibillec, J. et al. Transcriptome profile reveals AMPA receptor dysfunction in the hippocampus of the Rsk2-knockout mice, an animal model of Coffin–Lowry syndrome. Hum Genet 129, 255–269 (2011). https://doi.org/10.1007/s00439-010-0918-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-010-0918-0