Abstract
Mesoamerica, defined as the broad linguistic and cultural area from middle southern Mexico to Costa Rica, might have played a pivotal role during the colonization of the American continent. The Mesoamerican isthmus has constituted an important geographic barrier that has severely restricted gene flow between North and South America in pre-historical times. Although the Native American component has been already described in admixed Mexican populations, few studies have been carried out in native Mexican populations. In this study, we present mitochondrial DNA (mtDNA) sequence data for the first hypervariable region (HVR-I) in 477 unrelated individuals belonging to 11 different native populations from Mexico. Almost all of the Native Mexican mtDNAs could be classified into the four pan-Amerindian haplogroups (A2, B2, C1, and D1); only two of them could be allocated to the rare Native American lineage D4h3. Their haplogroup phylogenies are clearly star-like, as expected from relatively young populations that have experienced diverse episodes of genetic drift (e.g., extensive isolation, genetic drift, and founder effects) and posterior population expansions. In agreement with this observation, Native Mexican populations show a high degree of heterogeneity in their patterns of haplogroup frequencies. Haplogroup X2a was absent in our samples, supporting previous observations where this clade was only detected in the American northernmost areas. The search for identical sequences in the American continent shows that, although Native Mexican populations seem to show a closer relationship to North American populations, they cannot be related to a single geographical region within the continent. Finally, we did not find significant population structure in the maternal lineages when considering the four main and distinct linguistic groups represented in our Mexican samples (Oto-Manguean, Uto-Aztecan, Tarascan, and Mayan), suggesting that genetic divergence predates linguistic diversification in Mexico.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Although largely explored by many disciplines, the peopling of the Americas remains as an issue of intense debate. The tempo and mode in which modern humans colonized the New World through Beringia to the rest of the American continent remains unclear; with numerous hypotheses have been proposed in the last few decades (Campbell and Mithum 1979; Chakraborty and Weiss 1991; Greenberg et al. 1986; Laughlin 1988; Spencer et al. 1977; Szathmary et al. 1983; Wallace et al. 1985). However, a date of entry into the Americas roughly consistent with the archaeological record, has been placed around 15,000–17,000 years before present (ybp), followed by subsequent episodes of reduced population size, compatible with the current low genetic diversity observed in present day Native American populations (Mulligan et al. 2004).
Interest in the settlement of the New World has boosted genetic studies in Native American populations. These studies are based mainly on the analysis of the mitochondrial (mtDNA), a genome inherited exclusively throughout the maternal line. Most extant Native American mtDNAs, if not of recent admixture, descend from four main founding haplogroups, initially baptized as A, B, C, D (Torroni et al. 1993a) and further characterized as A2, B2, C1, and D1 (Achilli et al. 2008; Fagundes et al. 2008; Perego et al. 2009; Tamm et al. 2007); and five minor lineages, namely C4c, D2a, D3, D4h3, and X2a (Achilli et al. 2008; Bandelt et al. 2003; Brown et al. 1998; Fagundes et al. 2008; Schurr et al. 1990; Tamm et al. 2007; Torroni et al. 1992, 1993a). Based on mtDNA sequence data, Bonatto and Salzano (1997) proposed the hypothesis of a single and early migration that entered into the New World through Beringia where the population settled, expanded and diversified before the further colonization of the rest of the American continent. This “Out of Beringia” model, in contrast to the previous multiple migration scenarios for the Pleistocene peopling of the Americas, was compatible with recent studies supporting a single migration scenario. For instance, according to Tamm et al. (2007) and Fagundes et al. (2008), the ancestral population left from northeast Asia during the Last Glacial Maximum, defined as an interval centered on 21,000 ybp (Clark and Mix 2002), crossed through the Bering Strait bearing the five founder lineages and remained isolated long enough to generate the pan-American haplogroups A2, B2, C1, D1, and X2a (Achilli et al. 2008), which were distributed southward probably following a Pacific coastal route. This pattern of dispersion is indeed found in the actual distribution of Uto-Aztecan languages extending from the Western Plateau and Coast of California to Nicaragua in Central America (Miller 1983) and Oto-Manguean languages found from Middle America (Northeast Mexico) to Nicaragua and Honduras in Central America (Suaréz 1983). Recently, Perego et al. (2009) have reported the variation at two rare American mtDNA haplogroups D4h3 and X2a, based on complete genome sequencing of the mtDNA molecule, signaling for the first time the existence of two distinctive and almost concomitant Paleo-Indian migrations routes which would have occurred 15–17 kya from Beringia.
Specific geographic regions of the continent might have played a key role during the colonization of the Americas. One such region is the linguistic and cultural area called Mesoamerica (Campbell et al. 1986; Kirchhoff 1943; Pailes and Whitecotton 1995), which extends from middle southern Mexico to Guatemala, Belize, Honduras, El Salvador, Nicaragua, and Costa Rica, in which ancient native populations shared languages, traditions, customs, and history (López-Austin and Luján 2001). This evidence suggests that Mesoamerica represented a geographic bottleneck and possibly limited the gene flow between North and South America, and concentrated an invaluable source of diversity in that region. In fact, present day Mexican territories, which cover most of Mesoamerica, harbor one of the richest ethnic and linguistic diversities of the continent. Thus, Mexico has eleven linguistic families divided into 68 major linguistic groups which include 291 living languages and seven extinct ones (Inali 2007).
Despite the abundant genetic data on different Native American groups across the continent, the patterns of variability of Native Mexicans still remain unclear. Some prospective studies based on uniparental markers (Barrot et al. 2005; Buentello-Malo et al. 2003; Collins-Schramm et al. 2004; Rangel-Villalobos et al. 1999, 2000, 2001a, b) showed a relative genetic homogeneity of Mexican autochthonous populations. Other studies based on mixed urban samples (Mestizos) (Bonilla et al. 2005; Cerda-Flores et al. 2002a, b; Green et al. 2000) indicated that these groups are basically the result of admixture between Native Americans and Europeans but provided a poor idea about the genetic structure and diversity of autochthonous populations. Moreover, studies focused on ancient mtDNA (González-Oliver et al. 2001) have shown the presence of haplogroup A2, B2, and C1 (following the most recent nomenclature) mtDNA lineages in pre-Columbians; and analyses of extant mtDNA based on restriction enzymes have also shown the presence of the four major Amerindian haplogroups in Mexican samples (Penaloza-Espinosa et al. 2007). A deep genetic characterization of a larger number of Native Mexican populations might give insights, not only on the complex relationships between autochthonous groups inside Mexico, but also on the reconstruction of the human history of the Americas.
Here, we present mtDNA sequence data for 477 unrelated individuals belonging to eleven different native populations from Mexico, namely, Triqui, Tarahumara, Purépecha, Otomí, Mixtec, Nahua Xochimilco, Nahua Zitlala, Nahua Ixhuatlancillo, Nahua Necoxtla, Maya, and Pima. For the first time, an exhaustive analysis of maternal lineages in Native Mesoamerican populations is reported and the results are discussed within the context of continental genetic variation by constructing a mtDNA database with published reported sequences from populations from North, Central, and South America.
Materials and methods
Population sampling
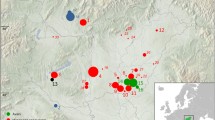
A total of 477 unrelated Mexican individuals from 11 populations were sampled with their appropriate informed consent. All individuals were native speakers with two generation-local unrelated ancestors. The geographic location of collected samples is shown in Fig. 1 and comprised 107 Triquis, 15 Tarahumaras, 34 Purépechas, 68 Otomíes, 35 Nahuas from Xochimilco, 14 Nahuas from Zitlala, 10 Nahuas from Ixhuatlancillo, 25 Nahuas from Necoxtla, 19 Mixtecs, 98 Pimas, and 52 Mayas. An additional sample of 23 Quechua individuals from Peru was included and used for population comparisons as part of the Native American database. All sequences are available at Supplementary Table S1.
Mitochondrial DNA genotyping
DNA was extracted from blood samples using standard phenol chloroform method (Sambrook et al. 1982) and FTA® extraction according to the supplier’s protocol. DNA from Pimas, Mayas, and Quechuas was directly provided from the laboratory of Judith R. Kidd and Kenneth K. Kidd (Yale University, New Haven, CT, USA) where lymphoblastoid cell lines are maintained. The mtDNA control region was amplified using primers L15996 and H408 (Vigilant et al. 1989). PCR products were subsequently purified with GFX PCR DNA Kit (Amersham Biosciences, GE Healthcare Bio-Sciences AB, Uppsala, Sweden). Both strands of the hypervariable region I (HVRI) were sequenced with the Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Carlsbad, CA, USA) using primers L15996 and H16401 (Vigilant et al. 1989). The sequence from positions 16024 to 16391 was determined (Anderson et al. 1981) for each individual. Length polymorphisms observed in the C-stretch from position 16184 to 16193 were not considered in those analyses involving population samples collected from the literature, due to lack of consistency when reporting variation at this region. In order to properly classify some sequences into haplogroups, additional markers in the mtDNA coding region were analyzed. The 9-bp tandem repeat (CCCCCTCTA) in the COII/tRNALys intergenic region, defining haplogroup B in Native Americans, was amplified using primers L8196 (5′-ACA GTT TCA TGC CCA TGG TC-3′) and H8297 (5′-ATG CTA AGT TAG CCT TAC AG-3′) and run into a 4% agarose gel (Comas et al. 2004). A diagnostic restriction enzyme (AluI for position 5176) was used to categorize haplogroup D, using the primers L5054 (5′-TAG GAT GAA TAA TAG CAG CTC TAC CG-3′), and H5189 (5′-GGG TGG ATG GAA TTA AGG GTG T-′3).
All the mtDNAs could be allocated to the Native American haplogroups A2, B2, C1, D1, and D4h3, with the exception of one Pima and one Quechua bearing West Eurasian haplogroups R1 and T, respectively. These two sequences were not considered for computational analysis. We follow Tamm et al. (2007), Fagundes et al. (2008), Achilli et al. (2008), and Perego et al. (2009) for the nomenclature of Native American clades. We are aware that HVRI alone is unable to distinguish Native American clades from their closest neighbors in northeast Asia; it is, however, commonly accepted that most of the variation belonging to haplogroups A, B, C, and D observed in Native Americans can be attributed to the American sub-lineages A2, B2, C1, and D1. This is particularly true for Mexico, where there is not known historical documentation supporting recent gene flow from Asian populations. We, therefore, use in what follows the latter nomenclature in the present study. Distinction of the minor Native American sub-clades, C4c, D2a, D3, D4h3, and X2a, as well as sub-lineages within A2, B2, C1, and D1, is also challenged by the limited resolution provided by the HVRI segment; some of these lineage, however, are characterized by diagnostic sites in this region (Achilli et al. 2008; Perego et al. 2009).
Data analysis
Intrapopulation genetic diversity parameters were computed using DnaSP software v4.0 (Rozas et al. 2003). The mean number of pairwise differences and the weighted intra-linage mean pairwise (WIMP) differences (Hurles et al. 2002) were calculated using the Arlequin program v3.1 (Excoffier et al. 2005).
A median joining network (Bandelt et al. 1995) was constructed using the Network 4.5.0.0 package (http://www.fluxus-engineering.com/) for each of the four main haplogroups (A2, B2, C1, and D1) found in Mexican populations. Positions 16024 to 16391 were considered and nucleotide position was given a weight as in Brandstätter et al. (2008) (Bandelt et al. 2006). The dating method employed (Saillard et al. 2000) is based on the average number of mutations accumulated from an ancestral sequence as a linear function of time and mutation rate. Values of ρ and σ were converted to age estimates using the most recent mutation rate available for the HVS-I segment of one transition per 18,845 years (in the sequence range 16090–16365) (Soares et al. 2009). This method was also performed with Network 4.5.0.0 program.
In order to compare the present Mexican results with other Native American populations, HVRI sequences from 52 Native American sample populations were collected from the literature (Supplementary Table S2). We removed from this database a total of 12 sequences belonging to non-Native American haplogroups (Achilli et al. 2008), considered to be the result of recent admixture with European and African individuals. For some analyses, population samples were grouped into major subcontinental areas: North (879 individuals from Alaska to southern Mexico), Central (234 individuals from Guatemala to Panama), and South America (1,633 individuals from Colombia southwards) (Table S2). For some analyses, Mexican samples were also considered as a separate category from North, Central, and South America. A spatial analysis of the molecular variance (SAMOVA) was also performed using the SAMOVA 1.0 program (Dupanloup et al. 2002) by presetting different numbers of population groups. This approach defines groups of populations that are geographically homogeneous and maximizes the proportion of total genetic variance due to differences between groups. Population genetic structure was tested through analysis of molecular variance (AMOVA) (Excoffier et al. 1992) using the Arlequin program v3.1 (Excoffier et al. 2005). Genetic relationships among Mexican and continental populations were analyzed by means of pairwise genetic differences and haplogroup absolute frequencies. The former were calculated using Arlequin program v3.1 (Excoffier et al. 2005) and represented in a Multidimensional Scaling plot (MDS) and the latter were used to construct a Correspondence Analysis (CA) plot using STATISTICA 7 package (http://www.statsoft.com).
In order to compare Mexican mtDNA sequences within the American framework, sequence variation within the range 16090 to 16362 (Anderson et al. 1981) was used for inter-population comparisons. Each mitochondrial sequence found in Mexico was compared with the dataset (Supplementary Table S3). To estimate the most likely source ancestral population (North, Central, or South America) of each of the Mexican mtDNA observed in our study, we followed a Bayesian approach: the probability of each of the subcontinental regions was computed as \( P_{{0{\text{s}}}} = {\frac{1}{n}}\sum\nolimits_{i = 1}^{n} {k_{i}{\frac{{p_{{i{\text{s}}}} }}{{p_{{i{\text{c}}}} }}}} \) where n is the number of Mexican sequences matching at least one mtDNA in the whole continental database; k i the number of times the sequence i is found in the Mexican sample; p is the frequency of the sequence i in the subcontinental region database; and p ic the frequency of the sequence i in the whole continental database. To provide confidence intervals for each of the estimations for the subcontinental regions, we also computed the standard deviations as \( SD(P_{{0{\text{s}}}} ) = \sqrt {{\frac{{p_{{0{\text{s}}}} (1 - p_{{0{\text{s}}}} )}}{n}}} \) (Mendizabal et al. 2008).
Results
Mitochondrial DNA diversity in Native Mexican populations
A total of 118 different sequences were found in the 477 Mexican individuals analyzed in the present study. The genetic diversity found in Mexican samples is summarized in Table 1. Three different estimates for the expected number of unique sequences in the different populations sampled are also provided (Egeland and Salas 2008) in Table 1, indicating that the samples sizes for the 12 ethnic groups analyzed in the present study cover reasonably well the expected variation at the HVRI segment. In addition, the sample coverage parameter (C; for definition see also Egeland and Salas 2008) also indicates that sample sizes represent in most of the cases more than 60% of the expected haplotypes in the populations; only the Quechua sample from Peru seems to represent only a very small proportion of the expected number of haplotypes in the population (~9%; data not shown). The results showed that Pimas (0.54 ± 0.05) and Triquis (0.55 ± 0.05) presented the lowest sequence diversity values despite their larger sample sizes. The average pairwise differences by population range from 1.69 to 7.42. Although most Mexican populations presented high sequence diversity and moderate mean pairwise differences, the WIMP differences value was low in all samples, suggesting that Mexican samples present a composition of distantly phylogenetic related haplogroups with low to moderate internal diversity.
All Mexican sequences were classified into the five Amerindian haplogroups A2, B2, C1, D1, and D4h3, according to Achilli et al. (2008) and Perego et al. (2009), with the exception of one Pima individual who presented a sequence probably belonging to the Eurasian haplogroup R1 as suggested by the presence of exact matches (16278T 16311C) to other sample populations where coding region information was also available (Macaulay et al. 1999). This sequence was excluded from subsequent analyses (Table 1). The most frequent haplogroup in Native Mexican populations was haplogroup A2 (50.5%), followed by C1 (28.5%), B2 (17.6%), D1 (2.7%), and D4h3 (0.4%) (Table 2). With respect to the pan-American haplogroups, we noted that the composition of Native Mexicans contrasts with the pattern reported by Salzano (2002) based on restriction endonuclease analysis, in which Mexican and Central American samples have higher frequencies of haplogroups A2 and B2 when compared with C1 and D1. This is especially relevant in the Pima and Tarahumara samples were C1 is very frequent compared to other Mexican samples. We did not find the minor haplogroup X2a, which has been reported at low frequencies in North American samples and absent in South America (see Perego et al. 2009 for a recent analysis of this haplogroup based on complete genome sequences). The highest expected frequency (F) of the unobserved X2a haplogroup in the population can be estimated assuming that the pattern of variability follows a Poisson distribution and using a confidence interval of 95%; then 1 − e−Fn = 0.95, where n represents sample size. Therefore, the maximum frequency of the X2a haplogroup in native Mexican populations given our sample size would be 0.6% (see Egeland and Salas 2008 for some caveats concerning this approach).
It is worth mentioning that the occurrences of two out of the three D4h3 mtDNAs observed in our samples were in one Mexican Tarahumara and one Peruvian Quechua, both living in the West side of the continent, as predicted by the results of (Perego et al. 2009), who postulated that this haplogroup spread into the Americas along the Pacific Coast. The third occurrence of D4h3 was in the Nahua from Ixhualtlancillo, a population living in the narrow region of southern Mexico, where it is also expected to find some members of this lineage (as it will probably occur in other studies on native populations living in other Mesoamerican countries).
The most common haplotypes are shared by almost all the populations. However, it is interesting to note that the pattern of haplotype frequencies varies significantly between populations (independently of their sample sizes). To give an example, haplotype 16111T 16223T 16290T 16319A 16362C makes up ~66% of the Triqui population, ~64% of the Nahuas from Zitlala, but it is ~10% of the Otomí sample and it is absent in Pima. The fact that these haplotypes are common in America and are phylogenetically related to each other (see below) indicates that genetic drift has played an important role in modulating their spatial distribution in Mexico.
In order to investigate the genetic relationship between the mtDNAs observed in our sample set, a median joining network was constructed for each of the most common mitochondrial haplogroups, namely A2, B2, C1, and D1. The four networks clearly have a star-like pattern and do not present any well-defined cluster (for an example of haplogroup A, see Supplementary Figure S1). The estimated ages for the four haplogroups were dated in a wide range of 18–31 kya. Given that recent studies by Tamm et al. (2007); Achilli et al. (2008) and Perego et al. (2009) based on complete genome analysis provided more recent ages for the four main Native American haplogroups; the ages obtained here probably represent an overestimate of the real haplogroup coalescent times likely due to the low resolution provided by the HVRI segment analyzed in the present study.
Genetic structure of Native Mexican populations
An AMOVA was performed in order to define the population structure of Mexican indigenous populations according to geographic and linguistic criteria (Table 3). When all Mexican populations were considered as a single group, 19.23% (p < 0.01) of the genetic variance was found between populations, showing a relevant genetic heterogeneity among Native Mexican populations. A significant proportion of the variance (19.17%; p < 0.01) was found between groups when populations were classified into northern (Tarahumara and Pima) and central-southern groups (Triqui, Purépecha, Otomí, Nahua, Mixtec, and Maya), meaning that significant mtDNA differences between the northern and central-southern Mexican populations exist, however no statistical differences were found between central (Purépecha, Otomí, and Nahua) and southern (Triqui, Mixtec, and Maya) groups (4.35%, p = 0.15). When the geographical location of the samples was taken into account in a SAMOVA analysis, the first grouping of populations was again Tarahumara and Pima versus the rest of populations, and subsequent partitions of the variance rendered only individual populations without a geographical structure (data not shown).
Four major linguistic families found in Mexico were represented in our studied populations: Oto-Manguean (Mixtec, Triqui and Otomí), Uto-Aztecan (Nahua populations, Tarahumara and Pima), Tarascan (Purépecha) and Mayan (Maya). A non-significant −0.45% of the variance was explained by these linguistic groups, suggesting a lack of correlation between mtDNA genetic diversity and linguistic classification.
Mexican populations within the American genetic landscape
In order to compare the mtDNA diversity found in Mexico to the rest of the American continent, a Correspondence Analysis (CA) based on haplogroup frequencies was performed considering the whole set of populations included in our dataset (Fig. 2). The two-dimension plot accounts for 53.49% of the total variation whereas 28.31% is explained by the first dimension. Although the distribution along the second dimension (25.18%) slightly suggests a North-South pattern, no particular clustering of populations can be clearly detected. It is noteworthy that most Mexican populations are closely related to other North and Central American populations, with the clear exception of Pima and Tarahumara which appear considerably separated from the rest. A similar pattern is shown when a MDS analysis is performed at a continental scale; that is, no clear clusters, and a few outliers caused by low sequence diversity, such as Pima and Triqui (Mexico), Ayoreo (Bolivia and northern Paraguay) and Emerillon (French Guiana) were observed (data not shown).
An AMOVA considering all Native American populations showed a high degree of heterogeneity between populations (16.86% of the genetic variance; p < 0.01). It is noteworthy that this mtDNA heterogeneity is of a similar magnitude to that observed considering only Mexican populations. In order to ascertain whether this heterogeneity was caused by geographical factors, three continental groups (North, Central and South) were considered (Table S2). The AMOVA showed that a non-significant 0.65% (p = 0.15) of the genetic variance was due to differences between these three sub-continental groups, whereas the genetic variance within the three groups remained highly significant (16.44%, p < 0.01), pointing to an important genetic heterogeneity within the sub-continental areas and a lack of continental structure. When Mexican samples were considered as a separate category from northern, central and southern groups, a mere 1.09% (p = 0.046) of the genetic variance was attributed to differences among groups, whereas the genetic heterogeneity within groups remained highly significant (16.08%, p < 0.01).
To obtain rough estimates for the relationship of Mexican mtDNA lineages at a finer scale, we searched exact matching sequences in our dataset. This analysis assumes that the frequencies found nowadays are representatives of extinct populations. Although this assumption is somehow unrealistic due to the dramatic genetic drift events experienced by most of the Native American populations, this proxy is still useful to better describe the geographical distribution of haplotype Mexican sequences within the American continent. All the Mexican lineages were compared to our dataset of 2,746 published Native American sequences (Table S2) divided into the three main aforementioned geographical regions (North, Central, and South). Only 21 Native Mexican lineages, representing 59% of the individuals, were also found in the America dataset (Table S3). The average of the proportions of Mexican sequences found in each geographical region can be used as a proxy to infer the relationship of these populations within the continent. Thus, the distribution of the Native Mexican sequences could be described as follows: 46.29% (SD 2.98%) to North, 15.86% (SD 2.19%) to Central and 37.83% (SD 2.89%) to South America. As expected, the mtDNA variability observed in Mexicans cannot be allocated to a single geographical region within the continent (due to the lack of a clear geographical structure in the Americas as shown, e.g., in the AMOVA analysis). However, the Mexican mtDNA pool shows a closer relationship to northern American populations.
Discussion
Native American populations show a unique pattern of genetic diversity as a result of different demographic processes—population bottlenecks, founder effects, genetic drift—involved in the colonization of the New World and the posterior European contact and African slave trade (Mulligan et al. 2004; Salas et al. 2004). Presently Mexico is mainly inhabited by two distinct population groups: (1) the so-called ‘Mestizos’, a term used in the country and widely accepted by Mexicans to designate individuals of recent admixed ancestry (although the term is meaningless from a genetic point of view; see comments in Salas et al. (2004)), and (2) the native indigenous Mexican groups defined by their language, cultural traits, ethnicity, oral history and customs as part of their cultural complexity. Despite ‘Mestizos’ representing nearly 95% of the Mexican population (INEGI 2007), the Native American component is highly prevalent in their gene pool. Thus, for instance, the study by Cerda-Flores et al. (2002a) based on short tandem repeats showed the admixed percentage contribution of ‘Mestizo’ Mexicans from northeastern locations as deriving from Spanish (54.99 ± 3.44), Amerindian (39.99 ± 2.57) and African (5.02 ± 2.82). The analysis of the mtDNA in ‘Mestizos’ has shown that their maternal component is an admixture of Native American (89.1%), European (5.4%), and African (4.5%) lineages (Green et al. 2000).
Besides the intrinsic interest of exploring the variability of Native American groups, their analysis is also pivotal to understand the genetic composition of admixed populations in the Americas. Our results show that the majority of the mtDNAs of autochthonous Mexican individuals can be allocated to one of the four most common Native American haplogroups (A2, B2, C1, D1) (Achilli et al. 2008). We did not observe the mtDNA contribution of African and European ancestry, with the exception of one Pima individual who presented a Eurasian lineage that could be attributed to recent gene flow. The haplogroup distribution observed is similar to that found in an ancient Maya population in which haplogroup A2 accounted for most of the samples (84%) followed by C1 (8%) and B2 (4%), although haplogroup D1 was not found (González-Oliver et al. 2001).
It is noteworthy that no traces of haplogroup X2a were observed in our native Mexican populations. In contrast with haplogroups A2 to D1, which have an East Asian origin (Torroni et al. 1993b), haplogroup X has its origins in West Eurasia, and its entrance into the Americas is more controversial. Haplogroup X2a is not present in Central and South Native American populations (Perego et al. 2009) and represents a clade that lacks close relatives in the Old Word, including Siberia (Reidla et al. 2003). Our results point to a geographical limit in Mesoamerica beyond which haplogroup X2a is not found. Fagundes et al. (2008) suggested that this haplogroup was part of the gene pool of a single Native American founding population and its low frequency is probably due to a failed expansion as a result of its geographic location in the expansion wave and/or its low initial frequency. The most recent study of Perego et al. (2009) suggested however that X2a could have moved from Beringia directly into the North American regions located East of the Rocky Mountains; the X2a expansion could have occurred in the Great Plains region, where the terminal part of the glacial corridor ended, and is in complete agreement with both the extent of diversity and distribution of X2a observed in modern Native American populations. The absence of X2a in our samples supports the idea that Mesoamerica played an important role during the colonization of the continent, restricting this haplogroup to the northernmost lands and shaping the diversity of the other founder haplogroups on their way down to Central and South America.
We have shown a relevant genetic heterogeneity of indigenous groups within Mexico, probably due to processes such as extensive isolation, genetic drift, and founder effects. These demographic events have modulated the distinctive patterns of frequencies of the four main Native American haplogroups in the different Mexican populations. Some populations, in particular the northernmost Tarahumara and Pima, showed larger genetic distances from the rest of the Mexican populations and different haplogroup spectra. When our mtDNA sequences were analyzed together within the continental landscape, no particular structure was detected for the Mexican populations, and sub-continental division does not account for the genetic differences among Native American populations. These results point to a common origin of Native American populations, including the Mexican groups, with extensive isolation and genetic drift, which might have produced an extremely high heterogeneity in their haplogroup patterns.
The Americas are one of the most linguistically diverse regions in the world comprising about 150 linguistic stocks (Nichols 1992). In addition, languages in the Americas show a striking and unparalleled diversity in their grammatical structures. Divergence across populations is caused by genetic drift; however, the differences diminish by extensive contact between populations, i.e., by genetic exchange. The principles governing genetic and linguistic patterns are of different nature and, therefore, it is not uncommon to observe differences between them (Comas et al. 2008). In the Mexican samples analyzed, four major linguistic families are represented (Oto-Manguean, Uto-Aztecan, Tarascan, and Mayan). However, we did not find a correlation between linguistic affiliation and their mtDNA gene pool when populations were grouped according to their linguistic families. The genetic differences between linguistic groups were not significant, which concomitantly translates into a high genetic heterogeneity within linguistic groups. This heterogeneity and the lack of correlation between mtDNA diversity and linguistic affiliation could be explained by the extensive isolation between Mexican indigenous populations. Nonetheless, Tarahumara and Pima show a certain degree of genetic differentiation from the other nine groups, which could be explained by cultural isolation beyond language; note that these are the only two populations analyzed that lay out of the Mesoamerican area. This fact contrasts with the Nahua populations, which despite being affiliated with Uto-Aztecan, the same linguistic family as Tarahumara and Pima, do belong to the cultural-geographical Mesoamerican group. Overall, the data suggest that the Mesoamerican geographic barrier could explain population differences between Native American groups due to genetic drift.
In summary, our findings suggest that the mtDNA diversity patterns in Native Mexicans have been mainly driven by genetic drift. Native Mexicans have preserved their native mtDNA background with almost no contribution from European and/or African populations. Although linguistic structure in Mexicans is highly stratified, we did not observe a correlation between linguistic and mtDNA patterns. Despite the fact that mtDNA represents just a small fraction of the human genome, it has been demonstrated once more to be useful to unravel demographic events in human populations. However, it would be worth exploring other genomic regions in Mexican DNA where natural selection could have played an important role in modulating their variation, providing the necessary adaptive skills in their way to the colonization of South America.
References
Achilli A, Perego UA, Bravi CM, Coble MD, Kong QP, Woodward SR, Salas A, Torroni A, Bandelt HJ (2008) The phylogeny of the four pan-American MtDNA haplogroups: implications for evolutionary and disease studies. PLoS ONE 3:e1764
Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465
Bandelt HJ, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753
Bandelt HJ, Herrnstadt C, Yao YG, Kong QP, Kivisild T, Rengo C, Scozzari R, Richards M, Villems R, Macaulay V, Howell N, Torroni A, Zhang YP (2003) Identification of Native American founder mtDNAs through the analysis of complete mtDNA sequences: some caveats. Ann Hum Genet 67:512–524
Bandelt HJ, Kong QP, Richards M, Macaulay V (2006) Estimation of mutation rates and coalescence times: some caveats. In: Bandelt HJ, Macaulay V, Richards M (eds) Human mitochondrial DNA and the evolution of Homo sapiens. Springer, Berlin, pp 47–90
Barrot C, Sánchez C, Ortega M, Gonzalez-Martin A, Brand-Casadevall C, Gorostiza A, Huguet E, Corbella J, Gene M (2005) Characterisation of three Amerindian populations from Hidalgo State (Mexico) by 15 STR-PCR polymorphisms. Int J Legal Med 119:111–115
Bonatto SL, Salzano FM (1997) A single and early migration for the peopling of the Americas supported by mitochondrial DNA sequence data. Proc Natl Acad Sci USA 94:1866–1871
Bonilla C, Gutierrez G, Parra EJ, Kline C, Shriver MD (2005) Admixture analysis of a rural population of the state of Guerrero, Mexico. Am J Phys Anthropol 128:861–869
Brandstätter A, Zimmermann B, Wagner J, Gobel T, Rock AW, Salas A, Carracedo Á, Parson W (2008) Timing and deciphering mitochondrial DNA macro-haplogroup R0 variability in Central Europe and Middle East. BMC Evol Biol 8:191
Brown MD, Hosseini SH, Torroni A, Bandelt HJ, Allen JC, Schurr TG, Scozzari R, Cruciani F, Wallace DC (1998) mtDNA haplogroup X: an ancient link between Europe/Western Asia and North America? Am J Hum Genet 63:1852–1861
Buentello-Malo L, Penaloza-Espinosa RI, Loeza F, Salamanca-Gómez F, Cerda-Flores RM (2003) Genetic structure of seven Mexican indigenous populations based on five polymarker loci. Am J Hum Biol 15:23–28
Campbell L, Mithum M (1979) The languages of Native America. University of Texas Press, Austin
Campbell L, Kaufman T, Smith-Stark T (1986) Mesoamerica as a linguistic area. Language 62:530–570
Cerda-Flores RM, Budowle B, Jin L, Barton SA, Deka R, Chakraborty R (2002a) Maximum likelihood estimates of admixture in Northeastern Mexico using 13 short tandem repeat loci. Am J Hum Biol 14:429–439
Cerda-Flores RM, Villalobos-Torres MC, Barrera-Saldana HA, Cortes-Prieto LM, Barajas LO, Rivas F, Carracedo A, Zhong Y, Barton SA, Chakraborty R (2002b) Genetic admixture in three Mexican Mestizo populations based on D1S80 and HLA-DQA1 loci. Am J Hum Biol 14:257–263
Chakraborty R, Weiss KM (1991) Genetic variation of the mitochondrial DNA genome in American Indians is at mutation-drift equilibrium. Am J Phys Anthropol 86:497–506
Clark P, Mix A (2002) Ice sheets and sea level of the Last Glacial maximum. Quat Sci Rev 21:1–7
Collins-Schramm HE, Chima B, Morii T, Wah K, Figueroa Y, Criswell LA, Hanson RL, Knowler WC, Silva G, Belmont JW, Seldin MF (2004) Mexican American ancestry-informative markers: examination of population structure and marker characteristics in European Americans, Mexican Americans, Amerindians and Asians. Hum Genet 114:263–271
Comas D, Plaza S, Wells RS, Yuldaseva N, Lao O, Calafell F, Bertranpetit J (2004) Admixture, migrations, and dispersals in Central Asia: evidence from maternal DNA lineages. Eur J Hum Genet 12:495–504
Comas D, Bosch E, Calafell F (2008) Human genetics and languages. In: Encyclopedia of life sciences. Wiley, Chichester. http://www.els.net/. doi:10.1002/9780470015902.a0020810
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581
Egeland T, Salas A (2008) Estimating haplotype frequency and coverage of databases. PLoS ONE 3:e3988
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Fagundes NJ, Kanitz R, Eckert R, Valls AC, Bogo MR, Salzano FM, Smith DG, Silva WA Jr, Zago MA, Ribeiro-dos-Santos AK, Santos SE, Petzl-Erler ML, Bonatto SL (2008) Mitochondrial population genomics supports a single pre-Clovis origin with a coastal route for the peopling of the Americas. Am J Hum Genet 82:583–592
González-Oliver A, Márquez-Morfin L, Jimenez JC, Torre-Blanco A (2001) Founding Amerindian mitochondrial DNA lineages in ancient Maya from Xcaret, Quintana Roo. Am J Phys Anthropol 116:230–235
Green LD, Derr JN, Knight A (2000) mtDNA affinities of the peoples of North-Central Mexico. Am J Hum Genet 66:989–998
Greenberg JH, Ii CGT, Zegura SL, Campbell L, Fox JA, Laughlin WS, EkJE Szathmary, Weiss KM, Woolford E (1986) The settlement of the Americas: a comparison of the linguistic, dental, and genetic evidence (and Comments and Reply). Curr Anthropol 27:477–497
Hurles ME, Nicholson J, Bosch E, Renfrew C, Sykes BC, Jobling MA (2002) Y chromosomal evidence for the origins of oceanic-speaking peoples. Genetics 160:289–303
Inali (2007) Catálogo de las lenguas indígenas nacionales: Variantes lingüísticas de México con sus autodenominaciones y referencias geoestadísticas, México
INEGI (2007) XII Censo General de Población y Vivienda 2000. Instituto Nacional de Estadística y Geografía, México
Kirchhoff P (1943) Mesoamérica: sus límites geográficos, composición étnica y caracteres culturales. Acta Americana 1:92–107 (Inter-American Society of Anthropology and Geography)
Laughlin WS (1988) From Ammassalik to Attu: 10, 000 years of divergent evolution. Objets et Mondes 25:141–148
López-Austin A, López-Luján L (2001) El pasado indígena. Fondo de Cultura Económica Editors, México
Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Bonne-Tamir B, Sykes B, Torroni A (1999) The emerging tree of West Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet 64:232–249
Mendizabal I, Sandoval K, Berniell-Lee G, Calafell F, Salas A, Martinez-Fuentes A, Comas D (2008) Genetic origin, admixture, and asymmetry in maternal and paternal human lineages in Cuba. BMC Evol Biol 8:213
Miller KW (1983) Uto-Aztecan languages. In Southwest. In: Alfonso Ortiz WCS (ed) Handbook of North American Indians, vol 10. Smithsonian Institution, Washington, pp 113–124
Mulligan CJ, Hunley K, Cole S, Long JC (2004) Population genetics, history, and health patterns in Native Americans. Annu Rev Genomics Hum Genet 5:295–315
Nichols J (1992) Linguistic diversity in space and time. The University of Chicago Press, Chicago
Pailes RA, Whitecotton JW (1995) The frontiers of Mesoamerica: Northern and Southern. In: Reyman JE (ed) The Gran Chichimeca: essays of the archaeology and ethnohistory of Northern Mesoamerica. Worldwide Archaeology series, Avebury
Penaloza-Espinosa RI, Arenas-Aranda D, Cerda-Flores RM, Buentello-Malo L, Gonzalez-Valencia G, Torres J, Alvarez B, Mendoza I, Flores M, Sandoval L, Loeza F, Ramos I, Munoz L, Salamanca F (2007) Characterization of mtDNA haplogroups in 14 Mexican indigenous populations. Hum Biol 79:313–320
Perego UA, Achilli A, Angerhofer N, Accetturo M, Pala M, Olivieri A, Kashani BH, Ritchie KH, Scozzari R, Kong QP, Myres NM, Salas A, Semino O, Bandelt HJ, Woodward SR, Torroni A (2009) Distinctive Paleo-Indian migration routes from Beringia marked by two rare mtDNA haplogroups. Curr Biol 19:1–8
Rangel-Villalobos H, Rivas F, Torres-Rodriguez M, Jaloma-Cruz AR, Gallegos-Arreola MP, López-Satow J, Cantu JM, Figuera LE (1999) Allele frequency distributions of six Amp-FLPS (D1S80, APO-B, VWA, TH01, CSF1PO and HPRTB) in a Mexican population. Forensic Sci Int 105:125–129
Rangel-Villalobos H, Rivas F, Sandoval L, Ibarra B, Garcia-Carvajal ZY, Cantu JM, Figuera LE (2000) Genetic variation among four Mexican populations (Huichol, Purepecha, Tarahumara, and Mestizo) revealed by two VNTRs and four STRs. Hum Biol 72:983–995
Rangel-Villalobos H, Jaloma-Cruz AR, Cerda-Aguilar L, Rios-Angulo CD, Mendoza-Carrera F, Patino-Garcia B, Sandoval-Ramirez L, Figuera-Villanueva LE (2001a) The genetic DNA trace in men: chromosome Y haplotypes in a Mexican population, analyzing 5 STRs. Rev Invest Clin 53:401–406
Rangel-Villalobos H, Jaloma-Cruz AR, Sandoval-Ramírez L, Velarde-Felix JS, Gallegos-Arreola MP, Figuera LE (2001b) Y-chromosome haplotypes for six short tandem repeats (STRs) in a Mexican population. Arch Med Res 32:232–237
Reidla M, Kivisild T, Metspalu E, Kaldma K, Tambets K, Tolk HV, Parik J, Loogvali EL, Derenko M, Malyarchuk B, Bermisheva M, Zhadanov S, Pennarun E, Gubina M, Golubenko M, Damba L, Fedorova S, Gusar V, Grechanina E, Mikerezi I, Moisan JP, Chaventre A, Khusnutdinova E, Osipova L, Stepanov V, Voevoda M, Achilli A, Rengo C, Rickards O, De Stefano GF, Papiha S, Beckman L, Janicijevic B, Rudan P, Anagnou N, Michalodimitrakis E, Koziel S, Usanga E, Geberhiwot T, Herrnstadt C, Howell N, Torroni A, Villems R (2003) Origin and diffusion of mtDNA haplogroup X. Am J Hum Genet 73:1178–1190
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Saillard J, Magalhaes PJ, Schwartz M, Rosenberg T, Norby S (2000) Mitochondrial DNA variant 11719G is a marker for the mtDNA haplogroup cluster HV. Hum Biol 72:1065–1068
Salas A, Richards M, Lareu MV, Scozzari R, Coppa A, Torroni A, Macaulay V, Carracedo A (2004) The African diaspora: mitochondrial DNA and the Atlantic slave trade. Am J Hum Genet 74:454–465
Salzano FM (2002) Molecular variability in Amerindians: widespread but uneven information. An Acad Bras Cienc 74:223–263
Sambrook J, Frisch EF, Maniatis TE (1982) Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory Press, New York
Schurr TG, Ballinger SW, Gan YY, Hodge JA, Merriwether DA, Lawrence DN, Knowler WC, Weiss KM, Wallace DC (1990) Amerindian mitochondrial DNAs have rare Asian mutations at high frequencies, suggesting they derived from four primary maternal lineages. Am J Hum Genet 46:613–623
Soares P, Ermini L, Thomson N, Mormina M, Rito T, Röhl A, Salas A, Oppenheimer S, Macaulay V, Richards M (2009) Correcting for purifying selection: an improved human mitochondrial molecular clock. Am J Hum Genet (in press)
Spencer RF, Jennings JD, Johnson E, King AR, Stern T, Stewart KM, Wallace WJ (1977) The Native Americans: ethnology and backgrounds of the North American Indians. Harper and Row Publishers, New York
Suaréz JA (1983) The Mesoamerican Indian languages (Cambridge Language Surveys). Cambridge University Press, New York
Szathmary EJ, Ferrell RE, Gershowitz H (1983) Genetic differentiation in Dogrib Indians: serum protein and erythrocyte enzyme variation. Am J Phys Anthropol 62:249–254
Tamm E, Kivisild T, Reidla M, Metspalu M, Smith DG, Mulligan CJ, Bravi CM, Rickards O, Martinez-Labarga C, Khusnutdinova EK, Fedorova SA, Golubenko MV, Stepanov VA, Gubina MA, Zhadanov SI, Ossipova LP, Damba L, Voevoda MI, Dipierri JE, Villems R, Malhi RS (2007) Beringian standstill and spread of Native American founders. PLoS ONE 2:e829
Torroni A, Schurr TG, Yang CC, Szathmary EJ, Williams RC, Schanfield MS, Troup GA, Knowler WC, Lawrence DN, Weiss KM et al (1992) Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics 130:153–162
Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, Larsen M, Smith DG, Vullo CM, Wallace DC (1993a) Asian affinities and continental radiation of the four founding Native American mtDNAs. Am J Hum Genet 53:563–590
Torroni A, Sukernik RI, Schurr TG, Starikorskaya YB, Cabell MF, Crawford MH, Comuzzie AG, Wallace DC (1993b) mtDNA variation of aboriginal Siberians reveals distinct genetic affinities with Native Americans. Am J Hum Genet 53:591–608
Vigilant L, Pennington R, Harpending H, Kocher TD, Wilson AC (1989) Mitochondrial DNA sequences in single hairs from a southern African population. Proc Natl Acad Sci USA 86:9350–9354
Wallace DC, Garrison K, Knowler WC (1985) Dramatic founder effects in Amerindian mitochondrial DNAs. Am J Phys Anthropol 68:149–155
Acknowledgments
We would like to thank Andrés Moreno-Estrada, Isabel Mendizabal, Chiara Batini (Unitat de Biologia Evolutiva, UPF), Michelle Gardner (Department of Molecular Neuroscience, Institute of Neurology, University College London) for their comments and help; Mònica Vallés (Unitat de Biologia Evolutiva, UPF), Stéphanie Plaza and Roger Anglada (Servei Genòmica, UPF) for their technical support; and Kenneth Kidd and Judith Kidd (Yale University) for kindly providing Pima, Mayan and Quechua samples. This research was supported by the Agencia Española de Cooperación Internacional (AECI, Programa de Cooperación Interuniversitaria e Investigación Científica entre España e Iberoamérica) (A/7694/07) and Direcció General de Recerca, Generalitat de Catalunya (2005SGR/00608). Karla Sandoval received a fellowship from the Consejo Nacional de Ciencia y Tecnología (CONACyT), México.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
439_2009_693_MOESM3_ESM.doc
Haplogroup frequencies classified by population in the dataset used for comparison to the Mexican sequences analyzed in the present study (DOC 328 kb)
439_2009_693_MOESM4_ESM.doc
Mitochondrial DNA sequences found in Mexico with their haplogroup classification and number of (identical) matches in the database used for comparison. Only positions from 16090 to 16362 were taken into account in the comparison database (DOC 60 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Sandoval, K., Buentello-Malo, L., Peñaloza-Espinosa, R. et al. Linguistic and maternal genetic diversity are not correlated in Native Mexicans. Hum Genet 126, 521–531 (2009). https://doi.org/10.1007/s00439-009-0693-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-009-0693-y