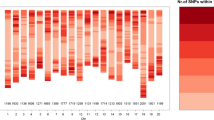
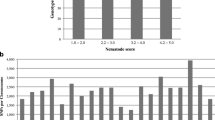
Abstract
Frogeye leaf spot, caused by the fungus Cercospora sojina, is a threat to soybeans in the southeastern and midwestern United States that can be controlled by crop genetic resistance. Limited genetic resistance to the disease has been reported, and only three sources of resistance have been used in modern soybean breeding. To discover novel sources and identify the genomic locations of resistance that could be used in soybean breeding, a GWAS was conducted using a panel of 329 soybean accessions selected to maximize genetic diversity. Accessions were phenotyped using a 1–5 visual rating and by using image analysis to count lesion number and measure the percent of leaf area diseased. Eight novel loci on eight chromosomes were identified for three traits utilizing the FarmCPU or BLINK models, of which a locus on chromosome 11 was highly significant across all model-trait combinations. KASP markers were designed using the SoySNP50K Beadchip and variant information from 65 of the accessions that have been sequenced to target SNPs in the gene model Glyma.11g230400, a LEUCINE-RICH REPEAT RECEPTOR-LIKE PROTEIN KINASE. The association of a KASP marker, GSM990, designed to detect a missense mutation in the gene was the most significant with all three traits in a genome-wide association, and the marker may be useful to select for resistance to frogeye leaf spot in soybean breeding.






Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files or are available from the corresponding author on reasonable request.
References
Afzal AJ, Wood AJ, Lightfoot DA (2008) Plant receptor-like serine threonine kinases: roles in signaling and plant defense. Mol Plant Microbe Interact 21:507–517. https://doi.org/10.1094/MPMI-21-5-0507
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664. https://doi.org/10.1101/gr.094052.109
Allen TW, Bradley CA, Sisson AJ et al (2017) Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2010 to 2014. Plant Health Progress 18:19–27. https://doi.org/10.1094/PHP-RS-16-0066
Athow KL, Probst AH (1952) The inheritance of resistance to frog-eye leaf spot on soybeans. Phytopathology 42:660–662
Athow K, Probst AH, Kartzman CP et al (1962) A newly identified physiological race of Cercospora sojina on soybean. Phytopathology 52:712–714
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265. https://doi.org/10.1093/bioinformatics/bth457
Bastien M, Sonah H, Belzile F (2014) Genome wide association mapping of Sclerotinia sclerotiorum resistance in soybean with a genotyping-by-sequencing approach. Plant Genome. https://doi.org/10.3835/plantgenome2013.10.0030
Bayer PE, Valliyodan B, Hu H et al (2022) Sequence the USDA core soybean collection reveals gene loss during domestication and breeding. Plant Genome 15:e20109. https://doi.org/10.1002/tpg2.20109
Bradbury PJ, Zhang Z, Kroon DE et al (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinform Appl 23:2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Bradley C, Allen T, Tenuta A, et al (2021) Soybean Disease Loss Estimates From the United States and Ontario, Canada 2020. https://cropprotectionnetwork.org/publications/soybean-disease-loss-estimates-from-the-united-states-and-ontario-canada-2020. Accessed 26 July 2022
Cingolani P, Platts A, Wang LL et al (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 6:80–92. https://doi.org/10.4161/fly.19695
Coser SM, Chowda Reddy RV, Zhang J et al (2017) Genetic architecture of charcoal rot (Macrophomina phaseolina) resistance in soybean revealed using a diverse panel. Front Plant Sci 8:1626. https://doi.org/10.3389/fpls.2017.01626
Cruz CD, Dorrance AE (2009) Characterization and survival of Cercospora sojina in Ohio. Plant Health Progress 10:17. https://doi.org/10.1094/PHP-2009-0512-03-RS
Danecek P, Auton A, Abecasis G et al (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158. https://doi.org/10.1093/bioinformatics/btr330
Dashiell KE, Akem CN (1991) Yield losses in soybeans from frogeye leaf spot caused by Cercospora sojina. Crop Prot 10:465–468. https://doi.org/10.1016/S0261-2194(91)80134-2
de Ronne M, Santhanam P, Cinget B et al (2021) Mapping of partial resistance to Phytophthora sojae in soybean PIs using whole-genome sequencing reveals a major QTL. Plant Genome. https://doi.org/10.1002/tpg2.20184
Díaz-Cruz G, Smith C, Wiebe K et al (2019) Applications of next generation sequencing for large scale pathogen diagnoses in soybean. Plant Dis 103:1075–1083. https://doi.org/10.1094/pdis-05-18-0905-re
Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JDG (1996) The tomato Cf-2 disease resistance locus comprises two function genes encoding leucine-rich repeat proteins. Cell 84:451–459. https://doi.org/10.1016/S0092-867(00)81290-8
Dixon MS, Hatzixanthis K, Jones DA, Harrison K, Jones JDG (1998) The tomato Cf-5 disease resistance locus gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10:1915–1925. https://doi.org/10.1105/tpc.10.11.1915
Fehr WR, Caviness CE, Burmood DT, Pennington JS (1971) Stage of development descriptions for soybeans, Glycine Max (L.) Merrill. Crop Sci 11:929–931. https://doi.org/10.2135/cropsci1971.0011183X001100060051x
Gizlice Z, Carter TE, Burton JW (1994) Genetic base for North American public soybean cultivars released between 1947 and 1988. Crop Sci 34:1143. https://doi.org/10.2135/cropsci1994.0011183X003400050001x
Grant D, Nelson RT, Cannon SB, Shoemaker RC (2009) SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. https://doi.org/10.1093/nar/gkp798
Grau CR, Dorrance AE, Bond J, Russin JS (2004) Fungal diseases. In: Boerma HR, Specht JE (eds) Soybeans: improvement, production, and uses. American Society of Agronomy, Crop Science Society of America and Soil Science Society of America, Madison, pp 679–763
Harrelson BC, Kemerait RC, Culbreath AK et al (2021) Assessment of quinone outside inhibitor sensitivity and frogeye leaf spot race of Cercospora sojina in Georgia soybean. Plant Dis 105:2946–2954. https://doi.org/10.1094/pdis-02-21-0236-re
Hartman GL, West ED, Herman TK (2011) Crops that feed the world 2. Soybean-worldwide production, use, and constraints caused by pathogens and pests. Food Secur 3:5–17. https://doi.org/10.1007/s12571-010-0108-x
Hoskins AJ (2011) Genetic mapping of soybean resistance genes to frogeye leaf spot in five Chinese plant introductions and efficiency of early generation selection for low phytate soybean lines. University of Georgia, Athens
Huang M, Liu X, Zhou Y et al (2018) BLINK: a package for the next level of genome-wide association studies with both individuals and markers in the millions. GigaScience 8:1–12. https://doi.org/10.1093/gigascience/giy154
Keim P, Olson TC, Shoemaker RC (1988) A rapid protocol for isolating soybean DNA. Soybean Genet Newslett 15:150–152
Koboldt DC, Steinberg KM, Larson DE et al (2013) The next-generation sequencing revolution and its impact on genomics. Cell 155:27–38. https://doi.org/10.1016/j.cell.2013.09.006
Liu X, Huang M, Fan B et al (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLOS Genet 12:e1005767. https://doi.org/10.1371/journal.pgen.1005767
McDonald SC, Buck J, Li Z (2022) Automated, image-based disease measurement for phenotyping resistance to soybean frogeye leaf spot. Plant Methods 18:103. https://doi.org/10.1186/s13007-022-00934-7
Melchers LE (1925) Diseases of cereal and forage crops in the United States in 1924. Plant Dis Rep 40:106–191
Mengistu A, Kurtzweil NC, Grau CR (2002) First report of frogeye leaf spot (Cercospora sojina) in Wisconsin. Plant Dis 86:1272. https://doi.org/10.1094/PDIS.2002.86.11.1272B
Mian MAR, Boerma HR, Phillips DV et al (1998) Performance of frogeye leaf spot-resistant and-susceptible near-isolines of soybean. Plant Dis 82:1017–1021
Mian MAR, Wang T, Phillips DV, Alvernaz J, Boerma HR (1999) Molecular mapping of the Rcs3 gene for resistance to frogeye leaf spot in soybean. Crop Sci 39:1687. https://doi.org/10.2135/cropsci1999.3961687x
Mian MAR, Missaoui AM, Walker DR, Phillips DV, Boerma HR (2008) Frogeye leaf spot of soybean: a review and proposed race designations for isolates of Cercospora sojina Hara. Crop Sci 48:14–24. https://doi.org/10.2135/cropsci2007.08.0432
Missaoui AM, Phillips DV, Boerma HR (2007) DNA marker analysis of ‘Davis’ soybean and its descendants for the Rcs3 gene conferring resistance to Cercospora sojina. Crop Sci 47:1263–1270. https://doi.org/10.2135/cropsci2006.07.0472
Mundt CC (2018) Pyramiding for resistance durability: theory and practice. Phytopathology 108:792–802. https://doi.org/10.1094/phyto-12-17-0426-rvw
Nascimento KJT, Debona D, Rezende D et al (2018) Changes in leaf gas exchange and chlorophyll a fluorescence on soybean plants supplied with silicon and infected by Cercospora sojina. J Phytopathol. https://doi.org/10.1111/jph.12757
Pace PF, Weaver DB, Ploper LD (1993) Additional genes for resistance to frogeye leaf spot race 5 in soybean. Crop Sci 33:1144–1145. https://doi.org/10.2135/cropsci1993.0011183X003300060006x
Pedersen WL, Leath S (2003) Pyramiding major genes for resistance to maintain residual effects. Annu Rev Phytopathol 26:369–378. https://doi.org/10.1146/annurev.py.26.090188.002101
Pham AT, Harris DK, Buck J et al (2015) Fine mapping and characterization of candidate genes that control resistance to Cercospora sojina K. Hara in two soybean germplasm accessions. PLoS ONE 10:e0126753. https://doi.org/10.1371/journal.pone.0126753
Phillips DV, Boerma HR (1981) Cercospora sojina race 5: a threat to soybeans in the Southeastern United States. Phytopathology 71:334–336
Phillips DV, Boerma HR (1982) Two genes for resistance to race 5 of Cercospora sojina in soybeans. Phytopathology 72:764–766
Probst AH, Athow KL, Laviolette FA (1965) Inheritance of resistance to race 2 of Cercospora sojina in soybeans. Crop Sci 5:332. https://doi.org/10.2135/cropsci1965.0011183X000500040013x
Ross JP (1968) Additional physiological races of Cercospora sojina on soybeans in North Carolina. Phytopathology 58:708–709
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. https://doi.org/10.1038/nmeth.2089
Singh T, Sinclair JB (1985) Histopathology of Cercospora sojina in soybean seeds. Phytopathology 75:185–189
Song WY, Wang GL, Chen LL et al (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270:1804–1806. https://doi.org/10.1126/science.270.5243.1804
Song Q, Hyten DL, Jia G et al (2015) Fingerprinting soybean germplasm and its utility in genomic research. G3 Genes Genom Genet. 5:1999–2006. https://doi.org/10.1534/g3.115.019000
Spearman C (1906) ‘Footrule’ for measuring correlation. Br J Psychol 1904–1920(2):89–108. https://doi.org/10.1111/j.2044-8295.1906.tb00174.x
Standish JR, Tomaso-Peterson M, Allen TW et al (2015) Occurrence of QoI fungicide resistance in Cercospora sojina from Mississippi Soybean. Plant Dis 99:1347–1352. https://doi.org/10.1094/PDIS-02-15-0157-RE
Sun M, Na C, Jing Y et al (2022) Genome-wide association analysis and gene mining of resistance to China Race 1 of frogeye leaf spot in soybean. Front Plant Sci. https://doi.org/10.3389/fpls.2022.867713
Sved JA (1971) Linkage disequilibrium of chromosome segments. Theor Popul Biol 2:125–141
Tibbs Cortes L, Zhang Z, Yu J (2021) Status and prospects of genome-wide association studies in plants. Plant Genome 14:1–17. https://doi.org/10.1002/tpg2.20077
Tran DT, Steketee CJ, Boehm JD et al (2019) Genome-wide association analysis pinpoints additional major genomic regions conferring resistance to soybean cyst nematode (Heterodera glycines Ichinohe). Front Plant Sci 10:1–13. https://doi.org/10.3389/fpls.2019.00401
Valliyodan B, Brown AV, Cannon SB, Nguyen H (2020) Genetic variation among 481 diverse soybean accessions. Sci Data. https://doi.org/10.1038/s41597-021-00834-w
Wang J, Zhang Z (2020) GAPIT Version 3: Boosting power and accuracy for genomic association and prediction 1 2. bioRxiv 2020.11.29.403170
Wang W, Chenn L, Fengler K et al (2021) A giant NLR gene confers broad-spectrum resistance to Phytophthora sojae in soybean. Nat Commun 12:6263. https://doi.org/10.1038/s41467-021-26554-8
Wise KA, Newman ME (2015) Frogeye leaf spot. In: Hartman GL, Rupe JC, Sikora EJ et al (eds) Compendium of soybean diseases, fifth. The American Phytopathological Society, Minnesota, pp 43–45
Yang XB, Feng F (2001) Ranges and diversity of soybean fungal diseases in North America. Phytopathology 91:769–775. https://doi.org/10.1094/PHYTO.2001.91.8.769
Yang W, Weaver DB (2001) Resistance to frogeye leaf spot in maturity groups VI and VII of soybean germplasm. Crop Sci 41:549–552. https://doi.org/10.2135/cropsci2001.412549x
Yorinori JT (1992) Management of foliar fungal diseases in soybean in Brazil. In: Coping LG (ed) Pest management in soybean. Springer, Dordrecht, pp 185–195
Zatybekov A, Abugalieva S, Didorenko S et al (2018) GWAS of a soybean breeding collection from South East and South Kazakhstan for resistance to fungal diseases. Vavilov J Genet Breed 22:536–543. https://doi.org/10.18699/VJ18.392
Zhang G, Pedersen DK, Phillips DV, Bradley CA (2012a) Sensitivity of Cercospora sojina isolates to quinone outside inhibitor fungicides. Crop Prot 40:63–68. https://doi.org/10.1016/j.cropro.2012.04.025
Zhang GR, Newman MA, Bradley CA (2012b) First report of the soybean frogeye leaf spot fungus (Cercospora sojina) resistant to quinone outside inhibitor fungicides in North America. Plant Dis 96:767–767. https://doi.org/10.1094/PDIS-10-11-0915-PDN
Zhang J, Singh A, Mueller DS, Singh AK (2015) Genome-wide association and epistasis studies unravel the genetic architecture of sudden death syndrome resistance in soybean. Plant J 84:1124–1136. https://doi.org/10.1111/tpj.13069
Acknowledgements
The authors wish to acknowledge the technical support provided by Carol Picard in conducting greenhouse disease assays and Nicole Bachleda and Tatyana Nienow in conducting marker assays. Thank you to Dr. Randall Nelson (USDA, retired) for providing seeds from the USDA Soybean Germplasm Collection used in this study.
Funding
Funding for this research was provided by the University of Georgia Research Foundation, Georgia Agricultural Commodity Commission for Soybeans, and Georgia Seed Development. This research was also supported by the funds allocated to the Georgia Agricultural Experiment Stations.
Author information
Authors and Affiliations
Contributions
ZL conceived the project, provided oversight of the experiments, interpreted the results, and edited the manuscript. SM conducted the experiments, performed the phenotyping, analyzed the data and drafted the manuscript. JB oversaw greenhouse phenotyping and edited the manuscript. QS selected the association panel and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no competing interests to declare.
Additional information
Communicated by Bing Yang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
438_2022_1986_MOESM1_ESM.eps
Supplementary file1 (EPS 357 KB) Scree plot of the first 10 principal coordinates (PC) from population structure analysis using the SoySNP50K Beadchip. The y-axis on the left shows the percent of total variation that each PC accounts for, and the y-axis on the right shows the variance for each PC
438_2022_1986_MOESM2_ESM.eps
Supplementary file2 (EPS 3510 KB) Maximum likelihood estimations of individual subpopulations for each accession in the association panel calculated in ADMIXTURE. K values ranged from 2 to 10 subpopulations
438_2022_1986_MOESM4_ESM.png
Supplementary file4 (PNG 81 KB) Genome-wide linkage disequilibrium (LD) decay plot for 329 soybean accessions based on 36,317 SNPs. r2 dropped to half of its maximum value at approximately 175 kb
438_2022_1986_MOESM5_ESM.eps
Supplementary file5 (EPS 2147 KB) Neighbor joining tree of the association panel. Sixty-five sequenced lines that were used to design KASP markers for variants in the chromosome 11 locus are colored in blue
438_2022_1986_MOESM6_ESM.eps
Supplementary file6 (EPS 1912 KB) Manhattan plots with SoySNP50K (black) and KASP (blue) markers for (a) FarmCPU model with visual rating trait, (b) FarmCPU model with lesion number trait, (c) FarmCPU model with percent of leaf area diseased trait, (d) BLINK model with visual rating trait, (e) BLINK model with lesion number trait, and (f) BLINK model with percent of leaf area diseased trait. Significant threshold for all was −log10(P) = 5.86
438_2022_1986_MOESM8_ESM.xlsx
Supplementary file8 (XLSX 25 KB) List of accessions in the association panel with their regions of origin, maturity groups, and BLUP values for visual rating, lesion number, and percent of leaf area diseased
438_2022_1986_MOESM9_ESM.xlsx
Supplementary file9 (XLSX 14 KB) List of 52 candidate genes and their annotations identified in the 8 haplotype blocks identified for their involvement in FLS resistance
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
McDonald, S.C., Buck, J., Song, Q. et al. Genome-wide association study reveals novel loci and a candidate gene for resistance to frogeye leaf spot (Cercospora sojina) in soybean. Mol Genet Genomics 298, 441–454 (2023). https://doi.org/10.1007/s00438-022-01986-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-022-01986-z