Abstract
Eucommia ulmoides (E. ulmoides) is a deciduous perennial tree belonging to the order Garryales, and is known as “living fossil” plant, along with ginkgo (Ginkgo biloba), metaspaca (Metasequoia glyptostroboides) and dove tree (Davidia involucrata Baill). However, the genetic diversity and population structure of E. ulmoides are still ambiguous nowdays. In this study, we re-sequenced the genomes of 12 E. ulmoides accessions from different major climatic geography regions in China to elucidate the genetic diversity, population structure and evolutionary pattern. By integration of phylogenetic analysis, principal component analysis and population structure analysis based on a number of high-quality SNPs, a total of 12 E. ulmoides accessions were clustered into four different groups. This result is consistent with their geographical location except for group samples from Shanghai and Hunan province. E. ulmoides accessions from Hunan province exhibited a closer genetic relationship with E. ulmoides accessions from Shanghai in China compared with other regions, which is also supported by the result of population structure analyses. Genetic diversity analysis further revealed that E. ulmoides samples in Shanghai and Hunan province were with higher genetic diversity than those in other regions in this study. In addition, we treated the E. ulmoides materials from Shanghai and Hunan province as group A, and the other materials from other places as group B, and then analyzed the evolutionary pattern of E. ulmoides. The result showed the significant differentiation (Fst = 0.1545) between group A and group B. Some candidate highly divergent genome regions were identified in group A by selective sweep analyses, and the function analysis of candidate genes in these regions showed that biological regulation processes could be correlated with the Eu-rubber biosynthesis. Notably, nine genes were identified from selective sweep regions. They were involved in the Eu-rubber biosynthesis and expressed in rubber containing tissues. The genetic diversity research and evolution model of E. ulmoides were preliminarily explored in this study, which laid the foundation for the protection of germplasm resources and the development and utilization of multipurpose germplasm resources in the future.



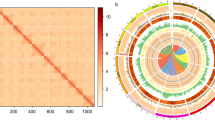
source population. (C) Principal component (PC) analysis, PC 1 against PC 2 and PC 3



Similar content being viewed by others
References
Abdel-Latif A, Osman G (2017) Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant Methods 13(1):1
Abdollahi Mandoulakani B, Yaniv E, Kalendar R, Raats D, Bariana HS, Bihamta MR, Schulman AH (2015) Development of IRAP- and REMAP-derived SCAR markers for marker-assisted selection of the stripe rust resistance gene Yr15 derived from wild emmer wheat. Theor Appl Genet 128(2):211–219
Ambreen H, Kumar S, Kumar A, Agarwal M, Jagannath A, Goel S (2018) Association mapping for important agronomic traits in safflower (Carthamus tinctorius L.) core collection using microsatellite markers. Front Plant Sci 9:402
Arzani A, Ashraf M (2016) Smart engineering of genetic resources for enhanced salinity tolerance in crop plants. Crit Rev Plant Sci 35(3):146–189
Baboo M, Dixit M, Sharma K, Saxena NS (2011) Mechanical and thermal characterization of cispolyisoprene and trans-polyisoprene blends. Polym Bull 66(5):661–672
Bamba T, Fukusaki E, Nakazawa Y, Kobayashi A (2002) In-situ chemical analyses of trans -polyisoprene by histochemical staining and Fourier transform infrared microspectroscopy in a rubber-producing plant, Eucommia ulmoides Oliver. Planta 215(6):934–939
Belcher B, Imang N, Achdiawan R (2004) Rattan, rubber, or oil palm: cultural and financial considerations for farmers in kalimantan. Econ Bot 58(1):S77–S87
Bhagyawant SS (2016) RAPD-SCAR markers: an interface tool for authentication of Traits. J Bioences Med 04(1):1–9
Chen R, Namimatsu S, Nakadozono Y, Bamba T, Nakazawa Y, Gyokusen K (2008) Efficient regeneration of Eucommia ulmoides from hypocotyl explant. Biol Plantarum 52(4):713–717
Cheng J, Long Y, Khan MA, Wei C, Fu S, Fu J (2015) Development and significance of RAPD-SCAR markers for the identification of Litchi chinensis Sonn by improved RAPD amplification and molecular cloning. Electron J Biotechn 18(1):35–39
Cherian S, Ryu SB, Cornish K (2019) Natural rubber biosynthesis in plants, the rubber transferase complex, and metabolic engineering progress and prospects. Plant Biotechnol J 17(11):2041–2061
Clément-Demange A, Priyadarshan PM, Hoa TT, Venkatachalam P (2007) Hevea Rubber Breeding and Genetics. Plant Breed Rev 29:177
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R (2011) The variant call format and VCFtools. Bioinformatics 27(15):2156–2158
Du H (2014) China Eucommia pictorial. China Forestry Publishing House, Beijing
Du H, Hu W, Yu R (2015) The report on development of China’s Eucommia rubber resources and industry (2014–2015). Social Sciences Academic Press, Beijing
Fu JJ, Mei ZQ, Tania M, Yang LQ, Cheng JL, Khan MA (2015) Development of RAPD-SCAR markers for different Ganoderma species authentication by improved RAPD amplification and molecular cloning. Genet Mol Res 14(2):5667–5676
Golkar P, Arzani A, Rezaei AM (2011) Genetic variation in safflower (Carthamus tinctorious L.) for seed quality-related traits and inter-simple sequence repeat (ISSR) markers. Int J Mol Sci 12(4):2664–2677
Golkar P, Nourbakhsh V (2019) Analysis of genetic diversity and population structure in Nigella sativa L. using agronomic traits and molecular markers (SRAP and SCoT). Ind Crop Prod 130:170–178
Hudson RR, Slatkin M, Maddison WP (1992) Estimation of gene flow from DNA sequence data. Genetics 132(2):583–589
Kumar J, Agrawal V (2017) Analysis of genetic diversity and population genetic structure in Simarouba glauca DC. (an important bio-energy crop) employing ISSR and SRAP markers. Ind Crop Prod 100:198–207
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549
Li H, Durbin R (2009) Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25(14):1754–1760
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The Sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079
Li Y, Wei H, Yang J, Du K, Li J, Zhang Y, Qiu T, Liu Z, Ren Y, Song L, Kang X (2020) High-quality de novo assembly of the Eucommia ulmoides haploid genome provides new insights into evolution and rubber biosynthesis. Hortic Res 7(1):183
Liu H, Lu Y, Wang J, Hu J, Wuyun T (2018) Genome-wide screening of long non-coding RNAs involved in rubber biosynthesis in Eucommia ulmoides. J Integr Plant Biol 60(11):1070–1082
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9):1297–1303
Najaphy A, Parchin RA, Farshadfar E (2012) Comparison of phenotypic and molecular characterizations of some important wheat cultivars and advanced breeding lines. Aust J Crop Sci 6(2):326–332
Quinn EM, Cormican P, Kenny EM, Hill M, Anney R, Gill M, Corvin AP, Morris DW (2013) Development of strategies for SNP detection in RNA-seq data: application to lymphoblastoid cell lines and evaluation using 1000 genomes data. PLoS ONE 8(3):e58815
Ramanatha Rao V, Hodgkin T (2002) Genetic diversity and conservation and utilization of plant genetic resources. Plant Cell Tissue Organ Cult 68(1):1–19
Reeves PA, Panella LW, Richards CM (2012) Retention of agronomically important variation in germplasm core collections: implications for allele mining. Theor Appl Genet 124(6):1155-1171
Rimbert H, Darrier B, Navarro J, Kitt J, Choulet F, Leveugle M, Duarte J, Rivière N, Eversole K, Le Gouis J, Davassi A, Balfourier F, Le Paslier M, Berard A, Brunel D, Feuillet C, Poncet C, Sourdille P, Paux E (2018) High throughput SNP discovery and genotyping in hexaploid wheat. PLoS ONE 13(1):e186329
Song K, Li L, Zhang G (2016) Coverage recommendation for genotyping analysis of highly heterologous species using next-generation sequencing technology. Sci Rep 6(1):35736–35736
Suzuki N, Uefuji H, Nishikawa T, Mukai Y, Yamashita A, Hattori M, Ogasawara N, Bamba T, Fukusaki E, Kobayashi A, Ogata Y, Sakurai N, Suzuki H, Shibata D, Nakazawa Y (2012) Construction and analysis of EST libraries of the trans-polyisoprene producing plant Eucommia ulmoides Oliver. Planta 236(5):1405–1417
Wang X, Luo G, Yang W, Li Y, Sun J, Zhan K, Liu D, Zhang A (2017) Genetic diversity, population structure and marker-trait associations for agronomic and grain traits in wild diploid wheat Triticum urartu. BMC Plant Biol 17(1):112
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7(2):256–276
Wright S (1951) The genetical structure of populations. Ann Eugen 15(4):323–354
Wuyun TN, Wang L, Liu H, Wang X, Zhang L, Bennetzen JL, Li T, Yang L, Liu P, Du L, Wang L, Huang M, Qing J, Zhu L, Bao W, Li H, Du Q, Zhu J, Yang H, Yang S, Liu H, Yue H, Hu J, Yu G, Tian Y, Liang F, Hu J, Wang D, Gao R, Li D, Du H (2018) The hardy rubber tree genome provides insights into the evolution of polyisoprene biosynthesis. Mol Plant 11(3):429–442
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Human Genet 88(1):76–82
Yao X, Deng J, Huang H (2012) Genetic diversity in Eucommia ulmoides (Eucommiaceae), an endangered traditional Chinese medicinal plant. Conserv Genet 13(6):1499–1507
Yu J, Wang Y, Peng L, Ru M, Liang ZS (2015) Genetic diversity and population structure of Eucommia ulmoides Oliver, an endangered medicinal plant in China. Genet Mol Res 14(1):2471–2483
Zhang Z, Dunwell JM, Zhang Y (2018) An integrated omics analysis reveals molecular mechanisms that are associated with differences in seed oil content between Glycine max and Brassica napus. BMC Plant Biol 18(1):328
Zhou H, Alexander D, Lange K (2011) A quasi-Newton acceleration for high-dimensional optimization algorithms. Stat Comput 21(2):261–273
Zhou Q, Zhou C, Zheng W, Mason AS, Fan S, Wu C, Fu D, Huang Y (2017) Genome-wide SNP markers based on SLAF-seq uncover breeding traces in rapeseed (Brassica napus L.). Front Plant Sci 8:648
Zhu M, Sun R (2018) Eucommia ulmoides Oliver: a potential feedstock for bioactive products. J Agr Food Chem 66(22):5433–5438
Acknowledgements
The authors sincerely thank the Research Institute of Non-timber Forestry, Chinese Academy of Forestry and Key Laboratory of Non-timber Forest Germplasm Enhancement & Utilization of State Forestry and Grassland Administration for the material, laboratory and data analysis guidance.
Funding
This work was financially supported by the Demonstration of Key Technologies and Industrial Applications of Eucommia Rubber (2019JZZY020223).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Bing Yang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Qing, J., Meng, YD., He, F. et al. Whole genome re-sequencing reveals the genetic diversity and evolutionary patterns of Eucommia ulmoides. Mol Genet Genomics 297, 485–494 (2022). https://doi.org/10.1007/s00438-022-01864-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-022-01864-8