Abstract
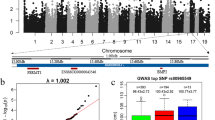
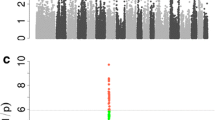
In China, sparerib is one of the most valuable parts of the pork carcass. As a result, candidate gene mining for number of ribs has become an interesting study focus. To examine the genetic basis for this major trait, we genotyped 596 individuals from an F2 Large White × Minzhu intercross pig population using the PorcineSNP60 Genotyping BeadChip. The genome-wide association study identified a locus for number of ribs in a 2.38-Mb region on Sus scrofa chromosome 7 (SSC7 of Sus scrofa genome assembly, Sscrofa10.2). We identified the top significant SNP ASGA0035536, which explained 16.51 % of the phenotypic variance. A previously reported candidate causal mutation (g.19034 A>C) in vertebrae development-associated gene VRTN explained 8.79 % of the phenotypic variation on number of ribs and had a much lower effect than ASGA0035536. Haplotype sharing analysis in F1 boars localized the rib number QTL to a 951-kb interval on SSC7. This interval encompassed 17 annotated genes in Sscrofa10.2, including the previously reported VRTN candidate gene. Of the 17 candidate genes, LTBP2, which encodes a latent transforming growth factor beta binding protein, was previously reported to indirectly regulate the activity of growth differentiation factor Gdf11, which has been shown to increase the number of ribs in knock-out mice. Thus, we propose LTBP2 as a good new candidate gene for number of ribs in the pig population. This finding advances our understanding of the genetic architecture of rib number in pigs.



Similar content being viewed by others
References
Borchers N, Reinsch N, Kalm E (2004) The number of ribs and vertebrae in a Pietrain cross: variation, heritability and effects on performance traits. J Anim Breed Genet 121:392–403
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Edwards DB, Ernst CW, Raney NE, Doumit ME, Hoge MD, Bates RO (2008) Quantitative trait locus mapping in an F2 Duroc × Pietrain resource population: II. Carcass and meat quality traits. J Anim Sci 86:254–266
Fan Y, Xing Y, Zhang Z, Ai H, Ouyang Z, Ouyang J, Yang M, Li P, Chen Y, Gao J, Li L, Huang L, Ren J (2013) A further look at porcine chromosome 7 reveals VRTN variants associated with vertebral number in Chinese and Western Pigs. PLoS One 8:e62534
Hayes AJ, Smith SM, Gibson MA, Melrose J (2011) Comparative immunolocalization of the elastin fiber-associated proteins fibrillin-1, LTBP-2, and MAGP-1 with components of the collagenous and proteoglycan matrix of the fetal human intervertebral disc. Spine (Phila Pa 1976) 36:E1365–E1372
Lee YJ, McPherron A, Choe S, Sakai Y, Chandraratna RA, Lee SJ, Oh SP (2010) Growth differentiation factor 11 signaling controls retinoic acid activity for axial vertebral development. Dev Biol 347:195–203
McPherron AC, Lawler AM, Lee SJ (1999) Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat Genet 22:260–264
Mikawa S, Hayashi T, Nii M, Shimanuki S, Morozumi T, Awata T (2005) Two quantitative trait loci on Sus scrofa chromosomes 1 and 7 affecting the number of vertebrae. J Anim Sci 83:2247–2254
Mikawa S, Sato S, Nii M, Morozumi T, Yoshioka G, Imaeda N, Yamaguchi T, Hayashi T, Awata T (2011) Identification of a second gene associated with variation in vertebral number in domestic pigs. BMC Genet 12:5
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215
Nezer C, Collette C, Moreau L, Brouwers B, Kim JJ, Giuffra E, Buys N, Andersson L, Georges M (2003) Haplotype sharing refines the location of an imprinted quantitative trait locus with major effect on muscle mass to a 250-kb chromosome segment containing the porcine IGF2 gene. Genetics 165:277–285
Ren DR, Ren J, Ruan GF, Guo YM, Wu LH, Yang GC, Zhou LH, Li L, Zhang ZY, Huang LS (2012) Mapping and fine mapping of quantitative trait loci for the number of vertebrae in a White Duroc × Chinese Erhualian intercross resource population. Anim Genet 43:545–551
Rifkin DB (2005) Latent transforming growth factor-β (TGF-β) binding proteins: orchestrators of TGF-β availability. J Biol Chem 280:7409–7412
Sato S, Oyamada Y, Atsuji K, Nade T, Sato S, Kobayashi E, Mitsuhashi T, Nirasawa K, Komatsuda A, Saito Y, Terai S, Hayashi T, Sugimoto Y (2003) Quantitative trait loci analysis for growth and carcass traits in a Meishan × Duroc F2 resource population. J Anim Sci 81:2938–2949
Shipley JM, Mecham RP, Maus E, Bonadio J, Rosenbloom J, McCarthy RT, Baumann ML, Frankfater C, Segade F, Shapiro SD (2000) Developmental expression of latent transforming growth factor β binding protein 2 and its requirement early in mouse development. Mol Cell Biol 20:4879–4887
Sun X, Essalmani R, Susan-Resiga D, Prat A, Seidah NG (2011) Latent transforming growth factor beta-binding proteins-2 and -3 inhibit the proprotein convertase 5/6A. J Biol Chem 286:29063–29073
Uemoto Y, Nagamine Y, Kobayashi E, Sato S, Tayama T, Suda Y, Shibata T, Suzuki K (2008) Quantitative trait loci analysis on Sus scrofa chromosome 7 for meat production, meat quality, and carcass traits within a Duroc purebred population. J Anim Sci 86:2833–2839
Zhang ZG, Li BD, Chen XH (1986) Pig breeds in China. Shanghai Scientific and Technical Publisher, Shang
Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, Buckler ES (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42:355–360
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Long-Chao Zhang declares that he has no conflict of interest. Jing-Wei Yue declares that she has no conflict of interest. Lei Pu declares that she has no conflict of interest. Li-Gang Wang declares that he has no conflict of interest. Xin Liu declares that she has no conflict of interest. Jing Liang declares that he has no conflict of interest. Hua Yan declares that she has no conflict of interest. Ke-Bin Zhao declares that he has no conflict of interest. Na Li declares that she has no conflict of interest. Hui-Bi Shi declares that he has no conflict of interest. Yue-Bo Zhang declares that he has no conflict of interest. Li-Xian Wang declares that he has no conflict of interest.
Additional information
Communicated by M. Fredholm.
L.-C. Zhang, J.-W. Yue and L. Pu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, LC., Yue, JW., Pu, L. et al. Genome-wide study refines the quantitative trait locus for number of ribs in a Large White × Minzhu intercross pig population and reveals a new candidate gene. Mol Genet Genomics 291, 1885–1890 (2016). https://doi.org/10.1007/s00438-016-1220-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-016-1220-1