Abstract
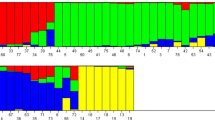
Knowledge of population structure and linkage disequilibrium among the worldwide collections of peppers currently classified as hot, mild, sweet and ornamental types is indispensable for applying association mapping and genomic selection to improve pepper. The current study aimed to resolve the genetic diversity and relatedness of Capsicum annuum germplasm by use of simple sequence repeat (SSR) loci across all chromosomes in samples collected in 2011 and 2012. The physical distance covered by the entire set of SSRs used was 2,265.9 Mb from the 3.48-Gb hot-pepper genome size. The model-based program STRUCTURE was used to infer five clusters, which was further confirmed by classical molecular-genetic diversity analysis. Mean heterozygosity of various loci was estimated to be 0.15. Linkage disequilibrium (LD) was used to identify 17 LD blocks across various chromosomes with sizes from 0.154 Kb to 126.28 Mb. CAMS-142 of chromosome 1 was significantly associated with both capsaicin (CA) and dihydrocapsaicin (DCA) levels. Further, CAMS-142 was located in an LD block of 98.18 Mb. CAMS-142 amplified bands of 244, 268, 283 and 326 bp. Alleles 268 and 283 bp had positive effects on both CA and DCA levels, with an average R 2 of 12.15 % (CA) and 12.3 % (DCA). Eight markers from seven different chromosomes were significantly associated with fruit weight, contributing an average effect of 15 %. CAMS-199, HpmsE082 and CAMS-190 are the three major quantitative trait loci located on chromosomes 8, 9, and 10, respectively, and were associated with fruit weight in samples from both years of the study. This research demonstrates the effectiveness of using genome-wide SSR-based markers to assess features of LD and genetic diversity within C. annuum.



Similar content being viewed by others
References
Aguilar-Melendez A, Morrell PL, Roose ML, Kim S-C (2009) Genetic diversity and structure in semiwild and domesticated chiles (Capsicum annuum; Solanaceae) from Mexico. Am J Bot 96(6):1190–1202
Arumugasundaram S, Ghosh M, Veerasamy S, Ramasamy Y (2011) Species discrimination, population structure and linkage disequilibrium in Eucalyptus camaldulensis and Eucalyptus tereticornis using SSR markers. PLoS One 6(12):e28252. doi:10.1371/journal.pone.0028252
Ben Chaim A, Paran I, Grube RC, Jahn M, van Wijk R, Peleman J (2001) QTL mapping of fruit-related traits in pepper (Capsicum annuum). Theor Appl Genet 102(6–7):1016–1028
Ben Chaim A, Borovsky Y, Rao GU, Tanyolac B, Paran I (2003) fs3.1: a major fruit shape QTL conserved in Capsicum. Genome 46(1):1–9
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57(1):289–300
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633–2635
Cong B, Barrero LS, Tanksley SD (2008) Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat Genet 40(6):800–804
Crossa J, Franco J (2004) Statistical methods for classifying genotypes. Euphytica 137(1):19–37
D’hoop B, Paulo MJ, Kowitwanich K, Sengers M, Visser RF, Eck H, Eeuwijk F (2010) Population structure and linkage disequilibrium unravelled in tetraploid potato. Theor Appl Genet 121(6):1151–1170
Emanuelli F, Lorenzi S, Grzeskowiak L, Catalano V, Stefanini M, Troggio M, Myles S, Martinez-Zapater J, Zyprian E, Moreira F, Grando M (2013) Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol 13(1):1–17
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14(8):2611–2620
Grandillo S, Ku HM, Tanksley SD (1999) Identifying the loci responsible for natural variation in fruit size and shape in tomato. Theor Appl Genet 99(6):978–987
Hamblin MT, Warburton ML, Buckler ES (2007) Empirical comparison of simple sequence repeats and single nucleotide polymorphisms in assessment of maize diversity and relatedness. PLoS One 2(12):e1367. doi:10.1371/journal.pone.0001367
Han K, Jeong H-J, Sung J, Keum Y, Cho M-C, Kim J-H, Kwon J-K, Kim B-D, Kang B-C (2013) Biosynthesis of capsinoid is controlled by the Pun1 locus in pepper. Mol Breed 31(3):537–548. doi:10.1007/s11032-012-9811-y
Hao C, Wang L, Ge H, Dong Y, Zhang X (2011) Genetic diversity and linkage disequilibrium in Chinese bread wheat (Triticum aestivum L.) revealed by SSR markers. PLoS One 6(2):e17279. doi:10.1371/journal.pone.0017279
Hernández-Verdugo S, Luna-Reyes R, Oyama K (2001) Genetic structure and differentiation of wild and domesticated populations of Capsicum annuum (Solanaceae) from Mexico. Plant Syst Evol 226(3–4):129–142. doi:10.1007/s006060170061
Hill TA, Ashrafi H, Reyes-Chin-Wo S, Yao J, Stoffel K, Truco M-J, Kozik A, Michelmore RW, Van Deynze A (2013) Characterization of Capsicum annuum genetic diversity and population structure based on parallel polymorphism discovery with a 30 K unigene pepper genechip. PLoS One 8(2):e56200. doi:10.1371/journal.pone.0056200
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23(14):1801–1806
Jin L, Lu Y, Xiao P, Sun M, Corke H, Bao J (2010) Genetic diversity and population structure of a diverse set of rice germplasm for association mapping. Theor Appl Genet 121(3):475–487
Liu S-R, Li W-Y, Long D, Hu C-G, Zhang J-Z (2013) Development and characterization of genomic and expressed SSRs in citrus by genome-wide analysis. PLoS One 8(10):e75149. doi:10.1371/journal.pone.0075149
Matsuoka Y, Mitchell SE, Kresovich S, Goodman M, Doebley J (2002) Microsatellites in Zea—variability, patterns of mutations, and use for evolutionary studies. Theor Appl Genet 104(2–3):436–450
Mazourek M, Pujar A, Borovsky Y, Paran I, Mueller L, Jahn MM (2009) A dynamic interface for capsaicinoid systems biology. Plant Physiol 150(4):1806–1821
Minamiyama Y, Tsuro M, Hirai M (2006) An SSR-based linkage map of Capsicum annuum. Mol Breed 18(2):157–169
Munos S, Ranc N, Botton E, Berard A, Rolland S, Duffe P, Carretero Y, Le Paslier MC, Delalande C, Bouzayen M, Brunel D, Causse M (2011) Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol 156(4):2244–2254
Ohashi J, Tokunaga K (2003) Power of genome-wide linkage disequilibrium testing by using microsatellite markers. J Hum Genet 48(9):487–491
Paran I (2003) Marker-assisted utilization of exotic germplam. In: Nguyen HT, Blum A (eds) Physiology and biotechnology integration for plant breeding. Marcel Dekker, New York
Paran I, Aftergoot E, Shifriss C (1998) Variation in Capsicum annuum revealed by RAPD and AFLP markers. Euphytica 99(3):167–173
Pickersgill (1997) Genetic resources and breeding of Capsicum spp. Euphytica 96:129–133
Portis E, Barchi L, Acquadro A, Macua JI, Lanteri S (2005) Genetic diversity assessment in cultivated cardoon by AFLP (amplified fragment length polymorphism) and microsatellite markers. Plant Breed 124(3):299–304
Powis TG, Gallaga Murrieta E, Lesure R, Lopez Bravo R, Grivetti L, Kucera H, Gaikwad NW (2013) Prehispanic use of chili peppers in Chiapas, Mexico. PLoS One 8(11):e79013. doi:10.1371/journal.pone.0079013
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Reddy UK, Almeida A, Abburi VL, Alaparthi SB, Unselt D, Hankins G, Park M, Choi D, Nimmakayala P (2014) Identification of gene-specific polymorphisms and association with capsaicin pathway metabolites in Capsicum annuum L. collections. PLoS One 9(1):e86393. doi:10.1371/journal.pone.0086393
Reyes-Valdés MH, Santacruz-Varela A, Martínez O, Simpson J, Hayano-Kanashiro C, Cortés-Romero C (2013) Analysis and optimization of bulk DNA sampling with binary scoring for germplasm characterization. PLoS One 8(11):e79936. doi:10.1371/journal.pone.0079936
Robbins MD, Sim S-C, Yang W, Van Deynze A, van der Knaap E, Joobeur T, Francis DM (2011) Mapping and linkage disequilibrium analysis with a genome-wide collection of SNPs that detect polymorphism in cultivated tomato. J Exp Bot 62(6):1831–1845
Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Mol Ecol Notes 4(1):137–138
Rousset F (2008) Genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 8(1):103–106
Schneider, Roessli D, Excoffier L (2000) Arlequin: a software for population genetics data analysis User manual
Sidak Z (1967) Rectangular confidence regions for the means of multivariate normal distributions. J Am Stat Assoc 62(318):626–633
Slatkin M (1985) Gene flow in natural populations. Annu Rev Ecol Syst 16:393–430
Sonah H, Deshmukh RK, Sharma A, Singh VP, Gupta DK, Gacche RN, Rana JC, Singh NK, Sharma TR (2011) Genome-wide distribution and organization of microsatellites in plants: an insight into marker development in Brachypodium. PLoS One 6(6):e21298. doi:10.1371/journal.pone.0021298
Sugita T, Semi Y, Sawada H, Utoyama Y, Hosomi Y, Yoshimoto E, Maehata Y, Fukuoka H, Nagata R, Ohyama A (2013) Development of simple sequence repeat markers and construction of a high-density linkage map of Capsicum annuum. Mol Breed 31(4):909–920
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Tenaillon MI, U’Ren J, Tenaillon O, Gaut BS (2004) Selection versus demography: a multilocus investigation of the domestication process in maize. Mol Biol Evol 21(7):1214–1225
Tomason Y, Nimmakayala P, Levi A, Reddy U (2013) Map-based molecular diversity, linkage disequilibrium and association mapping of fruit traits in melon. Mol Breed 31(4):829–841
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370
Yarnes SC, Ashrafi H, Reyes-Chin-Wo S, Hill TA, Stoffel KM, Van Deynze A, Gulick P (2012) Identification of QTLs for capsaicinoids, fruit quality, and plant architecture-related traits in an interspecific Capsicum RIL population. Genome 56(1):61–74
Yeh YR, Boyle T (1999) POPGENE Version 1.31. Microsoft Window-based Freeware for Population Genetic Analysis Quick User Guide
Yi G, Lee JM, Lee S, Choi D, Kim BD (2006) Exploitation of pepper EST-SSRs and an SSR-based linkage map. Theor Appl Genet 114(1):113–130
Yu J, Buckler ES (2006) Genetic association mapping and genome organization of maize. Curr Opin Biotechnol 17(2):155–160
Zhang J, Hao C, Ren Q, Chang X, Liu G, Jing R (2011) Association mapping of dynamic developmental plant height in common wheat. Planta 234(5):891–902
Zhao K, Aranzana MJ, Kim S, Lister C, Shindo C, Tang C, Toomajian C, Zheng H, Dean C, Marjoram P, Nordborg M (2007) An Arabidopsis example of association mapping in structured samples. PLoS Genet 3(1):e4
Zhu C, Gore M, Buckler ES, Yu J (2008) Status and prospects of association mapping in plants. Plant Gen 1(1):5–20
Acknowledgments
The study received funding from USDA-NIFA (2010-38821-21574), NSF-EPSCOR#09-570 - EPS-1003907, NIH Grant P20RR016477 to the West Virginia IDeA Network for Biomedical Research Funding and the Gus R. Douglass Institute (graduate research assistantship to V. Abburi).
Conflict of interest
We declare no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
P. Nimmakayala, V. L. Abburi and U. K. Reddy contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nimmakayala, P., Abburi, V.L., Abburi, L. et al. Linkage disequilibrium and population-structure analysis among Capsicum annuum L. cultivars for use in association mapping. Mol Genet Genomics 289, 513–521 (2014). https://doi.org/10.1007/s00438-014-0827-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-014-0827-3