Abstract
In the last years, we have seen the emergence of different tools that have changed the face of biology from a simple modeling level to a more systematic science. The transparent zebrafish embryo is one of the living models in which, after germline transformation with reporter protein-coding genes, specific fluorescent cell populations can be followed at single-cell resolution. The genetically modified embryos, larvae and adults, resulting from the transformation, are individuals in which time lapse analysis, digital imaging quantification, FACS sorting and next-generation sequencing can be performed in specific times and tissues. These multifaceted genetic and cellular approaches have permitted to dissect molecular interactions at the subcellular, intercellular, tissue and whole-animal level, thus allowing integration of cellular and developmental genetics with molecular imaging in the resulting frame of modern biology. In this review, we describe a new step in the zebrafish road to system biology, based on the use of transgenic biosensor animals expressing fluorescent proteins under the control of signaling pathway-responsive cis-elements. In particular, we provide here the rationale and details of this powerful tool, trying to focus on its huge potentialities in basic and applied research, while also discussing limits and potential technological evolutions of this approach.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Zebrafish (Danio rerio) is a small teleost that can be maintained in small aquaria. The main advantage of working with zebrafish is that it is an animal with all characteristics of vertebrates and an organism with external fertilization. These features, coupled with the fact that its embryo is semitransparent during early life stages, open the possibility to follow early development and organogenesis directly in vivo, without the need for invasive procedures. Moreover, zebrafish are particularly fertile and a healthy adult female can lay hundreds of eggs in a month. For these reasons, zebrafish is the first vertebrate model in which large-scale random mutagenesis could be undertaken (Driever et al. 1996; Haffter et al. 1996). The zebrafish genome has recently been sequenced and assembled, but its annotation is still ongoing. Moreover, until few years ago, genetic manipulation technology lacked a direct tool for targeting the zebrafish genome, but recently techniques have evolved through the use of TILLING (McCallum et al. 2000; Wienholds et al. 2003; Sood et al. 2006), zinc-finger nucleases (Meng et al. 2008; Doyon et al. 2008) and now TALEN mutagenesis, allowing conditional gene targeting (Sander et al. 2011; Huang et al. 2011). Being a vertebrate, zebrafish has immediately gained acceptance as a model organism in which human diseases can be phenocopied. In fact, many human diseases can be modeled in zebrafish and several laboratories all over the world are now generating fish with diseases that resemble, either at the molecular, cellular, etiological or histopathological level, human diseases. Several zebrafish models of cancer, metabolic, developmental, aging, degenerative, sensory organ and infectious diseases have been generated (reviewed in Lieschke and Currie 2007; Mione and Trede 2010; Kanther and Rawls 2010; Liu and Leach 2011; Goldsmith and Jobin 2012). This scenario makes zebrafish an ideal candidate for screening disease modifiers, genes, drugs or behavioral and environmental mechanisms able to modify specific phenotypes.
The knowledge of molecular and biochemical processes regulating cell proliferation, differentiation, survival, and death is the basis for understanding animal development, tissue homeostasis and tumor growth. Many embryonic and postnatal signaling pathways, which are often disrupted in cancer and diseases, appear to play a central role. Particularly important in regulating cell proliferation, stem cell maintenance and differentiation are bone morphogenetic proteins (BMPs), notch, wingless-related integration site (Wnt), sonic Hedgehog (Shh), fibroblast growth factor (FGF), signal transducer and activator of transcription (STAT), activator protein 1 (AP1) and cAMP-response element-binding protein (CREB) signaling (reviewed in Perrimon et al. 2012; Aaronson and Horvath 2002; Hess et al. 2004; Stork and Schmitt 2002). In fact, research at the tissue and cellular level is deeply oriented toward dissection of these pathways, as well as understanding their cross-talk and epistatic relations (Davidson 2006; Corson and Siggia 2012). As these pathways are relays of signal transduction, culminating with the activation of arrays of genes by specific set of transcription factors (TFs), it has been important in the last 30 years to understand the precise sequences and logic of gene activation. Typically, the analysis of genetic cascades culminates in dissecting cis-elements in the promoters of relevant target genes and the subsequent isolation of the TFs binding to these cis-elements. This approach leads to the isolation of relevant TFs in genetic cascades and the exact knowledge of the activator sequences in the promoters and enhancers of target genes (reviewed by Khalil et al. 2012). The direct and final proof that a specific TF (and thus the upstream genetic cascade) is involved in gene activation through a specific nucleotide sequence (cis-element) comes from the strong and tunable activation of synthetic constructs bearing multimerized copies of the cis-element bound to a minimal promoter of a reporter gene. In this way, a huge amount of data have been produced by transfection of a specific cell line together with the reporter and components of the signal transduction cascade. However, this approach might lack in vivo relevance, as co-transfection of a reporter plasmid together with its transduction and TFs leads, by default, to reporter signal. For this reason, a number of transgenic animal reporter lines have been developed, from mice to Drosophila and, recently, zebrafish. In this review, we will focus on the advantage of using zebrafish in this approach, to convince the reader that embryo and larval transparency, genetic and chemical manipulation, screening properties and transgenic manipulability, make zebrafish an ideal tool for analytical screenings and analysis of pathway reporters.
Development of signaling pathway reporters (SPRs)
The generation of reliable transgenic fish expressing in vivo reporter proteins under the control of signaling pathway responsive elements depends on four major critical issues: (a) choice of a suitable transgenesis method; (b) design and optimization of multimerized signaling responsive elements; (c) the use of a non-tissue specific and strong minimal promoter and (d) selection of an appropriate reporter protein (e.g. GFP, DsRed, mCherry, Kaede, Caerulean, YFP).
(a) Transgenesis
An ideal targeting vector should have a limited size and a high efficiency of germline transmission. Since the first successful attempt to introduce foreign DNA into the zebrafish germline (Stuart et al. 1988), considerable advances have been made in the field of gene transfer in fish. The initial method of transgenesis in zebrafish entailed the microinjection of linearized plasmids into fertilized eggs. However, this approach suffered from many drawbacks, including low rate of germline transmission and mosaic expression of the introduced transgene. Despite its limited yield and other minor issues, the first Wnt reporter line was established in the laboratory of Randall Moon in 2002 using this method (Dorsky et al. 2002).
An alternative transgenesis approach utilizes the I-SceI meganuclease, which was successfully introduced at first in Medaka (Oryzias latipes) and showed higher rates of germline transmission (Thermes et al. 2002). This method relies on the generation of a targeting sequence flanked by two recognition sites for the I-SceI enzyme, an intron-encoded homing endonuclease isolated from the yeast Saccharomyces cerevisiae. Co-injection of the vector with the I-SceI meganuclease significantly increases the rate of germline integration, thus improving the frequency of positive founder fish in F0. In zebrafish, the I-SceI mediated transgenesis protocol was shown to increase the transgenesis efficiency up to 45 % (Soroldoni et al. 2009). The combined use of a dual specificity phosphatase 6 (DUSP6) promoter-containing vector and the I-SceI meganuclease allowed the generation of the first transgenic reporter line specific for the FGF signaling pathway (Molina et al. 2007).
The development of the Tol2 technology has revolutionized transgenesis methods for aquatic vertebrates, such as zebrafish (Kawakami et al. 1998; Kawakami 2004; reviewed in Kawakami 2007). This method relies on the microinjection into fertilized eggs of a transposon-donor plasmid named Tol2, and a synthetic mRNA encoding the transposase, to catalyze DNA excision and recombination within the targeted genome. When considering the high integration frequency of the donor plasmid into the host genome during early stages of development, germline precursor cells carry at least one copy of the integrated plasmid and transmit it to the offspring with transgenesis rates that reach up to 70 % of the microinjected fish.
The Tol2 transgenesis method has been settled in most zebrafish laboratories worldwide and, in the past few years, many cell signaling reporter fish have been established through the application of this method (Schwend et al. 2010; Laux et al. 2011; Collery and Link 2011; Moro et al. 2012). Figure 1 summarizes the main steps to generate a zebrafish reporter line by the Tol2 strategy.
Generation of SPR zebrafish lines (Tol2 method). Pathway-specific responsive/regulatory elements (REs) can be obtained from literature data or predicted by in silico analysis (e.g. through UCSC Genome Browser) of conserved regions close to known target genes. To enhance the responsiveness of the planned reporter, REs can be multimerized, thus potentially recruiting more signal-dependent transcription factors (TF). Multimerized REs are cloned upstream of a Reporter gene coding for a fluorescent protein, provided with minimal promoter (MP) and polyA signal site (polyA), exploiting, for instance, the Gateway-based recombination cloning strategy. For Tol2 transgenesis, the complete transgene is cloned into a final destination vector with Tol2 integration sites. The construct is then co-injected into zebrafish zygotes along with the Tol2 transposase messenger RNA. The corresponding translated enzyme catalyzes the integration of the transgene into the zebrafish genome. F0 embryos are screened for their fluorescence (which may vary among individuals) and raised to adulthood (ca. 3 months). Putative founders are outcrossed with wild type fish and their F1 progeny screened for stable transgene integration and inheritance pattern
(b) Minimal regulatory elements
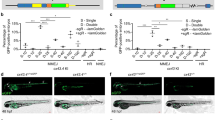
In most transgenic reporters, the responsiveness to a particular signaling pathway is achieved through the activation by TFs of regulatory elements located upstream to the coding sequences of a reporter protein (i.e. GFP). Pathway specificity and sequence information on these responsive elements can often be obtained through literature search, or identified by in silico analysis, taking advantage of ad hoc bioinformatics platforms. These sequence alignment tools are classically intended to compare promoter regions of co-expressed target genes, and are designed to detect evolutionarily conserved elements shared among different species (e.g. by phylogenetic footprinting) (Loots 2008). To enhance the reporter responsiveness, these regulatory DNA elements are synthesized in tandem repeats. This is, for instance, the case of the first zebrafish Wnt reporter line, which was created through the juxtaposition of four DNA regulatory elements specifically binding the TCF/Lef complex in Wnt responsive cells (Dorsky et al. 2002). The number of repeats and the extent of the binding consensus sequences of each repeat significantly affect the sensitivity of the reporter fish in terms of both expression levels and tissue specificity of reporter protein expression. In agreement with this hypothesis, a direct comparison between two Wnt reporter lines, the TOPdGFP (Dorsky et al. 2002) and the more recent TCFsiam Tg(7xTCFX.lasiam:GFP)ia4 (Moro et al. 2012), has been recently addressed. In the TOPdGFP transgenic line, four TCF/Lef responsive elements are located upstream of a cFos minimal promoter and the reporter protein coding sequence (d2GFP, destabilized GFP), while in the TCFsiam the reporter protein expression is driven by a multimerized array of seven TCF/Lef elements and a Xenopus leavis siamois gene minimal promoter. The TCFsiam line exhibits a higher mRNA expression level and a wider range of reporter protein-expressing domains in living embryos, which persists until late adulthood, supporting the idea that a higher number of responsive elements enhances the responsiveness of the reporter system to the binding TFs. However, while one could argue that differences in the minimal promoter sequence of the two Wnt lines, cFos and siamois, respectively, could account for the discrepancy in reporter activities, other explanations may derive from direct comparison of lines harboring the same minimal promoter, but a different number of responsive elements, as exemplified by the Notch reporter. The published Notch transgenic line, Tg(T2KTp1bglob:hmgb1-mCherry)jh11, is made of six copies of the promoter from the Epstein-Barr virus terminal protein (TP1) gene, each containing two Rbp-Jk binding sites, for a total of 12 Notch responsive elements (NREs), upstream to the rabbit beta-globin minimal promoter and the mCherry coding sequence (Parsons et al. 2009). We have recently generated two Notch reporter fish lines, one with 12 NREs and one with only 6 NREs upstream of the same minimal promoter and reporter protein coding sequence eGFP (unpublished data). When compared with the 12xNRE Notch reporter line, the 6xNRE transgenic fish show an identical domain of expression, but a less intense fluorescent signal in the Notch responsive cells (Fig. 2, compare a with a′). This observation suggests that cooperativity between responsive sequences affects the levels of reporter protein expression positively. At this point, it is important to stress that a higher responsiveness (potentially leading to signal saturation) does not translate to a better sensitivity of a specific reporter line in a particular experimental context. Indeed, the simultaneous use of different transgenic reporter lines with a different responsiveness for a given signaling pathway may be a better tool to address a specific biological question. To support this concept, very elegant work by the group of Richard Dorsky on Wnt pathway-dependent regulation of hypothalamic progenitors using two distinct reporter lines was recently published (Wang et al. 2012). This study took advantage of both stable and destabilized Wnt-responsive lines for two different purposes: the destabilized TOP:GFP reporter was used to rapidly assess conditionally induced Wnt inhibition in the hypothalamic region, while the long half life of GFP in the TCFSiam:GFP reporter was exploited to evaluate which hypothalamic lineages were derived from Wnt-responsive cells. A further example is shown in Fig. 2, where drug response of our 6xNRE reporter line is compared with that of 12xNRE transgenic fish. Both lines were treated with the Notch inhibitor DAPT and their fluorescence in vivo monitored at different time points (Fig. 2, compare a–c with a′–c′). After a 24-h treatment, the eGFP fluorescence is completely abolished in the 6xNRE line, while residual reporter protein is still present in the 12xNRE line, indicating the 6xNRE line as a faster biosensor to screen in vivo for Notch perturbing molecules. It is noteworthy that, compared to the fluorescent reporter, in situ hybridization analysis of the corresponding mRNAs shows faster dynamics (Fig. 2, compare d–f with d′–f′).
Number of responsive elements and reporter line sensitivity. Zebrafish embryos from two Notch-responsive lines, differing in the number of Notch-responsive elements (6x and 12xNRE), were treated with the Notch-inhibitor DAPT (100 µM) from 2 to 4 days post-fertilization. Controls were treated with a corresponding amount of carrier solution DMSO. a–c, a′–c′ In vivo analysis after 12 and 24 h of incubation shows faster decrease in EGFP reporter fluorescence in the 6xNRE line (a–c), as compared to 12xNRE fish (a′–c′). Complete suppression of EGFP fluorescence in 12xNRE embryos requires an extra day of incubation (not shown). d–f, d′–f′ Postmortem analysis by whole-mount in situ hybridization (WISH), with DIG-riboprobes specific for egfp mRNA, shows a rapid decrease of reporter messenger in both transgenic lines. WISH staining used far-red emitting fast blue, enhanced by alkaline-phosphatase-conjugated anti-DIG antibodies. All images were acquired with a Leica SP5 confocal microscope and processed by Volocity 6.0 software. All panels are dorsal views of the head region, anterior to the left. a, b, a′, b′, d, e, d′, e′ 60 h post-fertilization (hpf) stage; c, c′, f, f′ 72 hpf stage
(c) Minimal promoter for SPRs
Another key element that needs to be considered when designing SPR fish is the minimal promoter, which lies downstream of the signaling responsive elements. As previously pointed out (Barolo 2006), the use of minimal promoters taken from genes that are transcriptional targets of a specific signaling pathway is of great relevance when setting up a fish reporter for the chosen pathway. Both the TOPdGFP (Dorsky et al. 2002) and the TCFsiam lines (Moro et al. 2012), in which the minimal promoters used are cFos and siamois, respectively, show Wnt-dependent activation of the reporter transgene in several tissue contexts. On the other hand, however, very recent work by Shimizu et al. (2012) argued that in several Wnt reporter lines, including TOPdGFP and TCFSiam, the minimal promoters were derived from the promoters of natural genes, containing not only TATA, but also sequences derived from each gene promoter, which may be affected by non-specific signals. The authors suggested that the use of an artificial minimal promoter, such as the pGL4 vector-derived promoter called miniP, might increase the responsiveness and sensitivity of the reporter protein detection, reducing potentially unspecific signal. It should be emphasized that the choice of a particular minimal promoter does not follow a specific rule; therefore, when designing a SPR vector to be used in zebrafish, different minimal promoters might be tested in advance to select the one with maximal signal to noise ratio. Moreover, it should be clear that every newly designed SPR fish might suffer from limitations in terms of expression domains, as previously suggested (Barolo 2006).
(d) The reporter protein
The selection of a particular reporter protein is highly dependent on the type of research that is carried out. The choice of the best fluorophore should take into consideration a series of critical issues (reviewed in Miyawaki 2011; Shaner et al. 2007; Müller-Taubenberger and Anderson 2007), including type of color (e.g. based on the available optics or double-transgenic design), brightness, toxicity, tissue penetration, subcellular localization, availability of modified versions (e.g. convertible, toxic, stable or destabilized reporters) and so on. When addressing a biological question, dealing with rapid tissue or cell dynamics, a destabilized reporter protein, such as d2EGFP or VENUS-Pest are recommended. The application of such reporter systems has proven valuable, for instance, in detecting cell signaling changes during early stages of differentiation (Collery and Link 2011; Wang et al. 2012) or testing small chemical effects in short time ranges (Molina et al. 2009). However, the use of destabilized proteins often suffers from many drawbacks, including the low amount of detectable accumulating protein during a biological event. Therefore, especially for experimental tests involving analysis of cell signaling dynamics or cell lineage tracking over a long time range, the use of long half-life fluorescent proteins is suggested. We recently addressed the use of the Wnt reporter line, TCFsiam, to follow the cellular fates of neural crest-derived cells (NCdC) in the brain and trunk mesoderm. Using the stable GFP-expressing coding sequence, we were able to track the cellular division of NCdC in the CNS blood vessels for 9 h, a period far longer than the half life of the d2EGFP turnover (Moro et al. 2012). The use of a destabilized protein probably would not have allowed the detection of such cell division, as a consequence of low d2EGFP accumulation, given that its half life is 2 h.
Once all major variables (choice of targeting vector, number of responsive sequences, minimal promoter and reporter protein) have been addressed, the required components of the responsive transgene are routinely assembled into a final destination construct, for instance taking advantage of a Gateway-based Tol2 strategy (Kwan et al. 2007), and then microinjected into one cell-stage zebrafish embryos. It is generally assumed that Tol2 transgenesis leads to single gene integration into the host genome (Kawakami 2007). However, in practical experience, it is commonly observed that the generation of a new transgenic animal may be affected by multiple integration events occurring in the germline of microinjected fish. Considering that during multiple integration events, a transgene may be conditionally and spatially influenced at the transcriptional level by the genomic neighborhood (the so-called “positional effect”), some of the signaling reporter lines so far generated may display different patterns of expression in distinct isolated founders. This effect could hamper downstream applications and potentially generate misleading biological interpretations. It is of great relevance in such cases to carry out rigorous analysis and validation protocols to establish the accuracy of newly generated SPR lines.
Validation of SPR lines
How can we confirm that a given zebrafish transgenic line is a bona fide reporter for a specific signaling pathway? Here, we propose guidelines meant to provide consistency to a newly generated SPR line.
There are at least four major approaches to validate a SPR fish line: (a) comparison of multiple reporter lines, (b) literature review, (c) genetic analysis, and (d) pharmacological screening.
A first indication on reporter specificity can be obtained from simultaneous production and reciprocal comparison of multiple lines. The analysis of the progeny from several founder fish allows evaluation of whether a given transgene is consistently expressed in the same set of anatomical districts at equivalent time points. Differences in pattern and intensity among siblings are mostly due to position variegation effects or multiple insertions in the same founder (different alleles in the same carrier). On the other hand, possible discrepancies among progeny from different monoallelic founder fish are mainly ascribed to random positional effects due to the genomic context in which the single transgene integrated (Molina et al. 2007; Schwend et al. 2010; Moro et al. 2012). In our experience, these effects need to be attenuated by diluting the number of transgenic copies through subsequent generations, while keeping selected individuals with brightest and most shared positive signals, until the outcross will give a Mendelian proportion (50 % transgenic embryos) in progeny where sibs share similar expression levels. Generation and intercross of lines driving differently labeled reporters may further simplify these comparisons; exploiting dual color co-localization analysis of reporter expression in lines responsive to the same signaling pathway is a direct proof of identical or very similar pattern of expression and lack of a significant position effect.
Another basal set of evaluation criteria concerns the analysis of reporter expression compared to data available in current literature. The unequivocal identification of positive areas may require accurate morphological analysis, intercross of the reporter line with tissue-specific transgenic lines, or application of immunohistochemistry techniques. For instance, two-color fluorescent in situ hybridization, using reporter-specific probes (e.g. against GFP) in combination with other markers, may be particularly helpful in the validation process. Detected areas of reporter expression can thus be compared with known profiles of pathway-related target genes, or with anatomical districts whose development and function were previously linked to the activity of the pathway under investigation.
Finally, the last fundamental step in the validation of a biosensor line implies a series of functional studies, aimed to perturb a given pathway to verify whether the transgene is sufficiently sensitive in reporting signaling variations. These studies may apply genetic strategies, including over-expression of messengers encoding for pathway members, use of zebrafish mutants with defective signaling, or injection of antisense morpholino oligos. In some cases, these methodologies may be hindered by gene redundancy, non-specificity, hypomorphisms or other limitations, but, where effective, they appear convincing validations. In parallel, pharmacological approaches may be also adopted, such as treatments with agonistic/antagonistic drugs, aimed to perturb a given pathway at different levels of the signaling cascade. In the former type of validation methodology, based on the genetic approaches, the SPR line is generally tested with targeted antisense morpholino oligos against the specific molecules that directly drive or positively modulate the expression of the reporter transgene. As an example, the Tg(BmpRE:EGFP)pt510, which faithfully responds to BMP signaling activity, was successfully tested with SMAD5 and SMAD1 morpholinos (Laux et al. 2011). Knockdown of SMAD1 significantly dampens EGFP expression in many domains of the reporter line, confirming that the reporter expression is strictly regulated by nuclear TFs or cytoplasmic proteins that bind the responsive elements (in that case, ID1 protein responsive elements) in the nucleus.
However, given the transient activity and limited penetrance of morpholinos, the use of genetic mutants to test a SPR line is often favorable. In this case, the reporter line is first out-crossed with the genetic mutant, and then the offspring are intercrossed to generate homozygous mutants in the reporter background. If in the mutant the altered gene is a positive modulator of the signaling pathway displayed by the SPR, the homozygous fish mutants may show a complete abrogation of reporter expression in most tissue domains. This effect was partially shown in the Tg(Gli-d:mCherry) line, which display mCherry expression in Shh responsive domains (Schwend et al. 2010). The construct used to generate the line was tested in detour/gli1 (dtrts69), you-too/gli2 (yotty119), and slow-muscle-omitted/smu (smub641) mutants, and the lack of reporter expression allowed the authors to claim that their reporter line was a bona fide Shh reporter.
Because many genetic mutants are not viable beyond few days post-fertilization, the use of conditional systems, such as heat shock inducible lines, may be an ideal alternative approach to validate a SPR. For instance, the BMP signaling reporter Tg(BRE-AAVmlp:d2GFP)mw30 line was tested by performing intercrosses with Tg(hsp70 l:bmp2b)fr13 or with Tg(hsp70 l:nog3)fr14 line, which globally over-expresses zebrafish Bmp2b or the BMP inhibitor Noggin in response to heat-shock (Chocron et al. 2007), respectively. After heat shock, the double Tg(BRE-AAVmlp:d2GFP)mw30;Tg(hsp70 l:bmp2b)fr13 offspring displayed greatly increased expression of the d2GFP reporter, while in contrast Tg(BRE-AAVmlp:d2GFP)mw30;Tg(hsp70 l:nog3)fr14 double transgenic embryos showed reduced d2GFP levels. Similarly, we have recently tested the Tg(hsp70 l:dkk1-GFP) line (Stoick-Cooper et al. 2007) in the Wnt reporter background, showing that heat-shock induced expression of the Wnt antagonist Dkk1 is able to reduce the amount of Wnt-responsive reporter protein (Moro et al. 2012; Rampazzo et al. 2013).
In contrast to the above-mentioned genetic strategies, the pharmacological approach relies on testing the SPR fish with drugs that enhance or abrogate the activity of cytoplasmic proteins or more downstream nuclear TFs involved in the physiological responsiveness of the reporter transgene to endogenous ligands. For instance, the DAPT drug is known to block Notch signaling by inhibiting gamma secretase-dependent release of the intracellular domain of Notch receptor (Dovey et al. 2001). As shown in Fig. 2, treatment of Notch reporter fish with this compound leads to a decrease of Notch responsive activity, as demonstrated by the reduced amount of both reporter messengers and corresponding proteins. Table 1 provides a list of signaling pathways and related SPR transgenic lines produced so far, together with various drugs (pathway-specific activators or inhibitors) that have been used to validate each reporter line.
As mentioned before, these drug treatments are intended to be complementary and not exclusive methods for SPR validation. The combined use of all of the above-described strategies has indeed been shown to be the most reliable approach so far adopted to confirm, or exclude, the strength of a given SPR as a genuine reporter for a specific signaling pathway.
SPR lines: limits and applications
Zebrafish SPR lines are undoubtedly an invaluable resource for answering many unsolved biological questions regarding tissue communications and cell dynamics occurring in living vertebrates. Their application, however, also implies the knowledge of some possible limitations. First, these may include the incomplete recapitulation of a known signaling cascade in the corresponding SPR line, for instance in terms of number/type of anatomical districts expected to respond, at a given stage or condition, to the specific pathway. These discrepancies may be due to either expected or hard-to-predict genetic, epigenetic or non-genomic events, as well as to mere technical reasons. Some of these situations are clearly exemplified by the FGF reporter line (Molina et al. 2007). In these transgenic embryos, the expression domains of the reporter d2EGFP have not fully recapitulated all the anatomical regions (such as somites and tail bud) where a known FGF target, the gene dusp6, is normally activated. The authors discuss this observation, hypothesizing that the 10 Kb dusp6 promoter region, used to drive the transgene, might lack somitic and tail bud enhancers required for a complete reporter activation. In the same FGF reporter line, another discrepancy has been noted in the wider expression domain of the reporter messenger compared to the encoded d2EGFP. The authors argue that this difference might depend on the threshold level required to visualize the destabilized fluorescent protein, compared to WISH detection of reporter transcripts. Notably, the opposite may also occur, when stable fluorescent reporters display wider and more persistent expression compared to their messengers, which are usually more dynamically regulated (see, for instance, the Notch reporter behavior displayed in Fig. 2). When concerning another well-established reporter, the Hh-responsive line, Schwend et al. (2010) acknowledge that despite being the Hh-target gli1 domains plainly recapitulated by the reporter expression, some Hh-responding regions might not be represented, possibly due to local chromatin effects or other epigenetic factors. In this perspective, the examination of further reporter lines is proposed as a valid strategy to rule out this possibility.
In summary, several factors, including technical limitations in detecting reporter expression, choice of specific responsive elements or partial promoter regions, species-specific or stage-dependent responses, and non-canonical, non-genomic or epigenetic events, may all be taken into account when evaluating the activity of a given SPR transgenic line.
With all these issues in mind, the application of SPR lines to current research has met a large variety of needs so far, ranging from in vivo imaging of developmental and cellular dynamics to adult physiology and regeneration, tumor biology and pharmacological screening. The most obvious application of the existing zebrafish reporter lines is represented by the in vivo analysis of physiological pathway activation throughout the main steps of vertebrate embryonic development (Dorsky et al. 2002; Molina et al. 2007; Schwend et al. 2010; Gorelick and Halpern 2011; Moro et al. 2012). The peculiar optical clarity of zebrafish allows performing this task far beyond gastrulation, somitogenesis or late embryonic stages, reaching also larval or juvenile stages, especially when focusing on more superficial tissues, or exploiting unpigmented fish lines. Reporter analysis in adult individuals is also possible, even if generally focused on external structures or performed by postmortem organ dissection, followed by fluorescence microscopy and/or immunohistochemistry. A perfect example of reporter application in living adult fish is described in Moro et al. (2012), where amputated tail fins of 1-year old transgenic Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 fish are analyzed for Wnt/beta-catenin signaling activation during their epimorphic regeneration process.
Strictly connected with every SPR production is its validation, which, as described before, implies the use of genetic tools, to challenge the reporter behavior, as well as the test of compounds with agonistic/antagonistic effects on the pathway under investigation. Incidentally, these experiments have the additional advantage of validating SPR lines as suitable tools to spot, for instance, biologically significant differences among mutant alleles, or to set up large-scale screens for novel pathway-specific drugs, especially with potential therapeutic relevance. The use of zebrafish SPR lines for in vivo drug screening and validation, in particular, is expected to speed up and improve the whole methodological process, mainly based on the in vitro reporter systems, and commonly validated on model organisms by ex vivo or postmortem histological analyses (Chen et al. 2009). A recent example of such SPR utilization is represented by the GRIZLY assay, a chemical screening system based on glucocorticoid responsive zebrafish, which displays promising applications in stress research, environmental monitoring and drug discovery (Weger et al. 2012). Moreover, with novel SPR lines constantly becoming available, intercross between different transgenic reporters is nowadays feasible, allowing simultaneous analysis of multiple pathways at specific locations, conditions and time points (see, for instance Fig. 3a, b) and such applications are expected to improve our knowledge on interactions and hierarchical relationships (epistasis) among different signaling cascades activated during vertebrate development or key pathophysiological processes.
Examples of applications of zebrafish SPR lines. a, b Analysis of cephalic Wnt-reporter cells (red) in a tissue-specific transgenic background (in a vascular-specific kdrl:EGFP) or in combination with a Bmp-responsive system (in b Bmp:EGFP). White arrowheads (in b) indicate few cells co-expressing both Wnt- and Bmp-responsive fluorescent proteins. c Glioblastoma-derived (GBM) human cells, labeled with EGFP, are xeno-transplanted in the brain of a 1-week old Wnt-responsive zebrafish larva (0 dpt 0 days post-transplantation). Reporter expression (red) aids in locating transplanted cells close to the Wnt-responsive midbrain–hindbrain boundary (mhb). d In this microenvironment, at 1 dpt, a fraction of initially rounded GBM cells start to exhibit cellular projections (white arrows). e Ductal expression of the Notch reporter (red) in the pancreas (labeled in green by ptf1a:EGFP) of a control 1 month old (1 mpf) larva. f Disorganized appearance of the pancreatic tissue (green) in a 1 mpf larva expressing the oncogenic chimera Kras-EGFP. Notch-responsive ductal cells (red) appear completely dispersed. All figures are confocal Z-stack projections, in lateral view, anterior to the left, acquired with a Nikon C2 confocal microscope
With regard to SPR line application on disease modeling, it is worth mentioning some ongoing tumor studies carried out also by our group with various approaches, involving xeno-transplantation of human cancer cells into zebrafish larvae, and production of genetically engineered onco-fish lines. With these strategies, we aim to identify which signaling microenvironments can promote tumor growth, and which ones are instead able to re-program cancer cells into less aggressive phenotypes. To this end, we have exploited our SPR lines as hosting niches where human cancer cells could be grafted, targeting specific signaling-responsive anatomical regions (Fig. 3c). Xeno-transplanted cells could be monitored in vivo for several days, embedded in specific pathway-responsive backgrounds, and their phenotype morphologically examined (Fig. 3d) before proceeding with deeper molecular characterization and cell signaling analysis (Rampazzo et al. 2013). In parallel, our group has produced a number of zebrafish lines expressing the oncogene Kras in CNS and pancreas, to elicit endogenous medulloblastoma- and pancreatic adenocarcinoma-like tumor masses (Schiavone M and Rampazzo E, in preparation). These onco-fish have been crossed with different SPR lines to observe tumor behavior (onset, proliferation, metastasis, regression) in relation to each considered signaling cascade (Fig. 3e, f). With similar objectives, Santhakumar et al. (2012) have recently developed a zebrafish Hif (hypoxia inducible factor) signaling reporter, to analyze in vivo the hypoxia response in tumor-prone animals. To build up such reporter system, a Hif-responsive transgene, made by prolyl hydroxylase 3 (phd3) promoter/regulatory elements driving EGFP, was analyzed in vhl (Von Hippel-Lindau tumor suppressor) mutant zebrafish lines exposed to the carcinogen dimethylbenzanthracene (DMBA). Interestingly, DMBA-treated vhl fish showed a high incidence of hepatic and intestinal tumors, in a subset of which strong hypoxia-dependent phd3:EGFP expression could be noninvasively detected and characterized.
Taken together, these studies may thus represent a unique opportunity to dissect in real time in a vertebrate model the main steps of tumorigenesis while simultaneously inspecting a series of key signaling pathways activated at the whole-organism level.
SPR lines: further developments and conclusions
The generation of zebrafish SPR lines is just beginning, and several signaling pathways are still not represented by corresponding biosensors. These include, for instance, several hormone-dependent pathways, broad-spectrum cascades such as cAMP signaling, Jun/Fos/AP1, or recently characterized pathways, such as Hippo signaling. For the already available SPR lines, taking advantage of a plethora of available molecular tools, including destabilized (e.g. dEGFP), modifiable (e.g. Kaede, Dendra2) or phototoxic/cytotoxic (e.g. Killer Red, Nitroreductase) reporter proteins (Li et al. 1998; Ando et al. 2002; Gurskaya et al. 2006; Dempsey et al. 2012; Korzh et al. 2011; Curado et al. 2007) we can envisage a series of implementations aimed at improving biosensor performances in terms of a more dynamic response as well as the possibilities of cell lineage tracing or ablation. Moreover, spatial and temporal controls can be introduced in a given reporter system, exploiting, for instance, GAL4/UAS-coupled inducible expression, as already shown for the Notch reporter (Parsons et al. 2009). These systems can be further modulated by conditional and irreversible modifications, such as Cre/Lox recombination upon drug exposure (e.g. tamoxifen-regulated CreER), to drive the reporter in specific tissues or cell types, or regulated by temporary treatments, including heat-shock (e.g. Hsp70 promoter) (Stoick-Cooper et al. 2007; Hans et al. 2009). For more advanced SPR imaging technologies, further improvements are expected to arise in parallel with the set-up of large-scale automated small molecule screens, as well as through the combination of imaging and system biology, as recently demonstrated by the authors of Vibe-Z, software developed to map expression data in 3D zebrafish brain atlases (Ronneberger et al. 2012). In this perspective, Walker et al. (2012) recently described an interesting ad hoc system for Automated Reporter Quantification in vivo (ARQiv), based on a microplate reader designed to detect changes in fluorescent reporters expressed in living zebrafish from embryonic to larval and juvenile stages. This versatile platform, designed to quantify reporter signals at a high-throughput scale at different time points, should be able, for instance, to monitor disease progression or drug action kinetics.
In conclusion, we picture a rapid spread of zebrafish-based SPRs among the scientific community, especially in view of the fact that these biosensors fully meet the 4R principle of experimental animal use (Banks 1995). Undeniably, zebrafish SPRs allow replacement of amniote models with more economical anamniote organisms; moreover, zebrafish SPRs can be refined toward early life stages (embryos, larvae and juveniles). Finally, zebrafish SPRs significantly reduce the number of experimental animals by means of repeatable live imaging, in this way fully exploiting the extraordinary properties of manipulability and optical accessibility of this vertebrate model, without affecting a responsible use of animals in research protocol.
References
Aaronson DS, Horvath CM (2002) A road map for those who don’t know JAK-STAT. Science 296:1653–1655
Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A (2002) An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci USA 99:12651–12656
Banks RE (1995) The 4th R of research. Contemp Top Lab Anim Sci 34:50–51
Barolo S (2006) Transgenic Wnt/TCF pathway reporters: all you need is Lef? Oncogene 25:7505–7511
Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, Wei S, Hao W, Kilgore J, Williams NS, Roth MG, Amatruda JF, Chen C, Lum L (2009) Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol 5:100–107
Collery RF, Link BA (2011) Dynamic smad-mediated BMP signaling revealed through transgenic zebrafish. Dev Dyn 240:712–722
Corson F, Siggia ED (2012) Geometry, epistasis, and developmental patterning. Proc Natl Acad Sci USA 109:5568–5575
Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, Stainier DY (2007) Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn 236:1025–1035
Davidson EH (2006) The regulatory genome:gene regulatory networks in development and evolution, New edn. Academic, Oxford
Dempsey WP, Fraser SE, Pantazis P (2012) PhOTO zebrafish: a transgenic resource for in vivo lineage tracing during development and regeneration. PLoS ONE 7:e32888
Dodge ME, Moon J, Tuladhar R, Lu J, Jacob LS, Zhang LS, Shi H, Wang X, Moro E, Mongera A, Argenton F, Karner CM, Carroll TJ, Chen C, Amatruda JF, Lum L (2012) Diverse chemical scaffolds support direct inhibition of the membrane-bound O-acyltransferase porcupine. J Biol Chem 287:23246–23254
Dorsky RI, Sheldahl LC, Moon RT (2002) A transgenic Lef1/beta-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev Biol 241:229–237
Dovey HF, John V, Anderson JP, Chen LZ, de Saint Andrieu P, Fang LY, Freedman SB, Folmer B, Goldbach E, Holsztynska EJ, Hu KL, Johnson-Wood KL, Kennedy SL, Kholodenko D, Knops JE, Latimer LH, Lee M, Liao Z, Lieberburg IM, Motter RN, Mutter LC, Nietz J, Quinn KP, Sacchi KL, Seubert PA, Shopp GM, Thorsett ED, Tung JS, Wu J, Yang S, Yin CT, Schenk DB, May PC, Altstiel LD, Bender MH, Boggs LN, Britton TC, Clemens JC, Czilli DL, Dieckman-McGinty DK, Droste JJ, Fuson KS, Gitter BD, Hyslop PA, Johnstone EM, Li WY, Little SP, Mabry TE, Miller FD, Audia JE (2001) Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J Neurochem 76:173–181
Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL (2008) Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol 26:702–708
Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C (1996) A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123:37–46
Goldsmith JR, Jobin C (2012) Think small: zebrafish as a model system of human pathology. J Biomed Biotechnol 2012:817341
Gorelick DA, Halpern ME (2011) Visualization of estrogen receptor transcriptional activation in zebrafish. Endocrinology 152:2690–2703
Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA (2006) Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol 24:461–465
Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, Vogelsang E, Beuchle D, Schach U, Fabian C, Nusslein-Volhard C (1996) The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123:1–36
Hans S, Kaslin J, Freudenreich D, Brand M (2009) Temporally-controlled site-specific recombination in zebrafish. PLoS ONE 4:e4640
Hess J, Angel P, Schorpp-Kistner M (2004) AP-1 subunits: quarrel and harmony among siblings. J Cell Sci 117:5965–5973
Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B (2011) Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol 29:699–700
Kanther M, Rawls JF (2010) Host–microbe interactions in the developing zebrafish. Curr Opin Immunol 22:10–19
Kawakami K (2004) Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol 77:201–222
Kawakami K (2007) Tol2: a versatile gene transfer vector in vertebrates. Genome Biol 8(Suppl 1):S7
Kawakami K, Koga A, Hori H, Shima A (1998) Excision of the tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene 225:17–22
Khalil AS, Lu TK, Bashor CJ, Ramirez CL, Pyenson NC, Joung JK, Collins JJ (2012) A synthetic biology framework for programming eukaryotic transcription functions. Cell 150:647–658
Korzh V, Teh C, Kondrychyn I, Chudakov DM, Lukyanov S (2011) Visualizing compound transgenic zebrafish in development: a tale of green fluorescent protein and KillerRed. Zebrafish 8:23–29
Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB (2007) The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 236:3088–3099
Laux DW, Febbo JA, Roman BL (2011) Dynamic analysis of BMP-responsive smad activity in live zebrafish embryos. Dev Dyn 240:682–694
Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR (1998) Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem 273:34970–34975
Lieschke GJ, Currie PD (2007) Animal models of human disease: zebrafish swim into view. Nat Rev Genet 8:353–367
Liu S, Leach SD (2011) Zebrafish models for cancer. Annu Rev Pathol 6:71–93
Loots GG (2008) Genomic identification of regulatory elements by evolutionary sequence comparison and functional analysis. Adv Genet 61:269–293
McCallum CM, Comai L, Greene EA, Henikoff S (2000) Targeted screening for induced mutations. Nat Biotechnol 18:455–457
Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA (2008) Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol 26:695–701
Mione MC, Trede NS (2010) The zebrafish as a model for cancer. Dis Model Mech 3:517–523
Miyawaki A (2011) Proteins on the move: insights gained from fluorescent protein technologies. Nat Rev 12:656–668
Molina GA, Watkins SC, Tsang M (2007) Generation of FGF reporter transgenic zebrafish and their utility in chemical screens. BMC Dev Biol 7:62
Molina G, Vogt A, Bakan A, Dai W, Queiroz de Oliveira P, Znosko W, Smithgall TE, Bahar I, Lazo JS, Day BW, Tsang M (2009) Zebrafish chemical screening reveals an inhibitor of Dusp6 that expands cardiac cell lineages. Nat Chem Biol 5:680–687
Moro E, Ozhan-Kizil G, Mongera A, Beis D, Wierzbicki C, Young RM, Bournele D, Domenichini A, Valdivia LE, Lum L, Chen C, Amatruda JF, Tiso N, Weidinger G, Argenton F (2012) In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev Biol 366:327–340
Müller-Taubenberger A, Anderson KI (2007) Recent advances using green and red fluorescent protein variants. Appl Microbiol Biotechnol 77:1–12
Parsons MJ, Pisharath H, Yusuff S, Moore JC, Siekmann AF, Lawson N, Leach SD (2009) Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech Dev 126:898–912
Perrimon N, Pitsouli C, Shilo BZ (2012) Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb Perspect Biol 4:a005975
Rampazzo E, Persano L, Pistollato F, Moro E, Frasson C, Porazzi P, Della Puppa A, Bresolin S, Battilana G, Indraccolo S, Te Kronnie G, Argenton F, Tiso N, Basso G (2013) Wnt activation promotes neuronal differentiation of Glioblastoma. Cell Death Dis 4:e500
Ronneberger O, Liu K, Rath M, Ruebeta D, Mueller T, Skibbe H, Drayer B, Schmidt T, Filippi A, Nitschke R, Brox T, Burkhardt H, Driever W (2012) ViBE-Z: a framework for 3D virtual colocalization analysis in zebrafish larval brains. Nat Methods 9:735–742
Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR (2011) Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol 29:697–698
Santhakumar K, Judson EC, Elks PM, McKee S, Elworthy S, van Rooijen E, Walmsley SS, Renshaw SA, Cross SS, van Eeden FJ (2012) A zebrafish model to study and therapeutically manipulate hypoxia signaling in tumorigenesis. Cancer Res 72:4017–4027
Schwend T, Loucks EJ, Ahlgren SC (2010) Visualization of Gli activity in craniofacial tissues of hedgehog-pathway reporter transgenic zebrafish. PLoS ONE 5:e14396
Shaner NC, Patterson GH, Davidson MW (2007) Advances in fluorescent protein technology. J Cell Sci 120:4247–4260
Shimizu N, Kawakami K, Ishitani T (2012) Visualization and exploration of Tcf/Lef function using a highly responsive Wnt/beta-catenin signaling-reporter transgenic zebrafish. Dev Biol 370:71–85
Sood R, English MA, Jones M, Mullikin J, Wang DM, Anderson M, Wu D, Chandrasekharappa SC, Yu J, Zhang J, Paul Liu P (2006) Methods for reverse genetic screening in zebrafish by resequencing and TILLING. Methods 39:220–227
Soroldoni D, Hogan BM, Oates AC (2009) Simple and efficient transgenesis with meganuclease constructs in zebrafish. Methods Mol Biol 546:117–130
Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT (2007) Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development 134:479–489
Stork PJ, Schmitt JM (2002) Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol 12:258–266
Stuart GW, McMurray JV, Westerfield M (1988) Replication, integration and stable germ-line transmission of foreign sequences injected into early zebrafish embryos. Development 103:403–412
Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS (2002) I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev 118:91–98
Valdivia LE, Young RM, Hawkins TA, Stickney HL, Cavodeassi F, Schwarz Q, Pullin LM, Villegas R, Moro E, Argenton F, Allende ML, Wilson SW (2011) Lef1-dependent Wnt/beta-catenin signalling drives the proliferative engine that maintains tissue homeostasis during lateral line development. Development 138:3931–3941
Walker SL, Ariga J, Mathias JR, Coothankandaswamy V, Xie X, Distel M, Koster RW, Parsons MJ, Bhalla KN, Saxena MT, Mumm JS (2012) Automated reporter quantification in vivo: high-throughput screening method for reporter-based assays in zebrafish. PLoS ONE 7:e29916
Wang X, Kopinke D, Lin J, McPherson AD, Duncan RN, Otsuna H, Moro E, Hoshijima K, Grunwald DJ, Argenton F, Chien CB, Murtaugh LC, Dorsky RI (2012) Wnt signaling regulates postembryonic hypothalamic progenitor differentiation. Dev Cell 23:624–636
Weger BD, Weger M, Nusser M, Brenner-Weiss G, Dickmeis T (2012) A chemical screening system for glucocorticoid stress hormone signaling in an intact vertebrate. ACS Chem Biol 7:1178–1183
Wienholds E, van Eeden F, Kosters M, Mudde J, Plasterk RH, Cuppen E (2003) Efficient target-selected mutagenesis in zebrafish. Genome Res 13:2700–2707
Wilkinson RN, Pouget C, Gering M, Russell AJ, Davies SG, Kimelman D, Patient R (2009) Hedgehog and Bmp polarize hematopoietic stem cell emergence in the zebrafish dorsal aorta. Dev Cell 16:909–916
Acknowledgments
The authors apologize to all colleagues whose work could not be cited due to space constraints. The University of Pennsylvania Postdoctoral Editors Association is kindly acknowledged for manuscript editing. The work of FA, NT, NF, MA and MM is supported by the European Union Grant ZF-HEALTH CT-2010-242048. AV and AC are supported by the Cariparo Project “An in vivo reporter platform for cancer studies and drugs screening” to FA; MS is supported by the AIRC Project IG 10274 to FA. ER is supported by a fellowship from Bando Soci AIRC 2012. EM is supported by the Italian Ministry of Health (GR-2008-1139743). IZ is a recipient fellow of the project GR-2008-1139743. OE is supported by a Wallonie-Bruxelles International fellowship. The University of Padova Grant CPDA089044 and the Italian Ministry of Health Grant 2309484 support the work of NT.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Graw.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Moro, E., Vettori, A., Porazzi, P. et al. Generation and application of signaling pathway reporter lines in zebrafish. Mol Genet Genomics 288, 231–242 (2013). https://doi.org/10.1007/s00438-013-0750-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-013-0750-z