Abstract
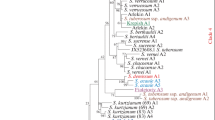
In planta the enzymatic activity of apoplastic and vacuolar invertases is controlled by inhibitory proteins. Although these invertase inhibitors (apoplastic and vacuolar forms) have been implicated as contributing to resistance to cold-induced sweetening (CIS) in tubers of potato (Solanum tuberosum L.), there is a lack of information on the structure and allelic diversity of the apoplastic invertase inhibitor genes. We have PCR-isolated and sequenced the alleles of the apoplastic invertase inhibitor gene (Stinh1) from three tetraploid potato genotypes: 1021/1 (a genotype with very high tolerance to CIS), ‘Karaka’ and ‘Summer Delight’ (two cultivars that are highly susceptible to CIS). In total, five alleles were identified in these genotypes, of which four (Stinh1-c, Stinh1-d, Stinh1-e, Stinh1-f) were novel. An analysis of allele diversity was conducted by incorporating previously published sequences of apoplastic invertase inhibitors from potato. Eight alleles were assessed for sequence polymorphism in the two exons and the single hypervariable intron. Contrary to the hypervariable intron, only 65 single nucleotide polymorphisms were observed in the exons, of which 42 confer amino acid substitutions. Phylogenetic analysis of amino acid sequences indicates that the alleles of the invertase inhibitor are highly conserved amongst members of the Solanaceae family.



Similar content being viewed by others
References
Anderson JAD, Genet RA, Triggs CM, McKenzie MJ (2005) The crisping performance and resistance to cold induced sweetening in different potato lines during long term cool storage. Acta Hort 670:151–156
Baldwin SJ, Dodds KG, Auvray B, Genet RA, Macknight RC, Jacobs JME (2011) Association mapping of cold-induced sweetening in potato using historical phenotypic data. Ann Appl Biol 158:248–256
Bendtsen JD, Jensen LJ, Blom N, von Heijne G, Brunak S (2004) Feature based prediction of non-classical and leaderless protein secretion. Protein Eng Des Select 17:349–356
Bernatzky R, Tanksley SD (1986) Genetics of actin related sequences in tomato. Theor Appl Genet 72:314–321
Birney E, Clamp M, Durbin R (2004) GeneWise and Genomewise. Genome Res 14:988–995
Bracho GE, Whitaker JR (1990) Characteristics of the inhibition of potato (Solanum tuberosum) invertase by an endogenous proteinaceous inhibitor in potatoes. Plant Physiol 92:381–385
Breathnach R, Chambon P (1981) Organization and expression of eukaryotic split genes coding for proteins. Ann Rev Biochem 50:349–383
Brummell DA, Chen RKY, Harris JH, Zhang H, Hamiaux C, Kralicek AV, McKenzie MJ (2011) Induction of vacuolar invertase inhibitor mRNA in potato tubers contributes to cold-induced sweetening resistance and includes spliced hybrid mRNA variants. J Exp Bot 62:3519–3534
Chen X, Salamini F, Gebhardt C (2001) A potato molecular-function map for carbohydrate metabolism and transport. Theor Appl Genet 102:284–295
Cheng WH, Taliercio EW, Chourey PS (1996) The Míniature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell 8:971–983
Cheng S-H, Liu J, Xie C-H, Song B-T, Li J-C (2007) Role of tobacco vacuolar invertase regulated by patatin promoter in potato tuber resistance to cold-sweetening. Chin J Agric Biotechnol 4:81–86
Dale MFB, Bradshaw JE (2003) Progress in improving processing attributes in potato. Trends Plant Sci 8:310–312
Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A (2011) Geneious v5.4. Available from http://www.geneious.com/
Greiner S, Krausgrill S, Rausch T (1998) Cloning of a tobacco apoplasmic invertase inhibitor: proof of function of the recombinant protein and expression analysis during plant development. Plant Physiol 116:733–742
Greiner S, Rausch T, Sonnewald U, Herbers K (1999) Ectopic expression of a tobacco invertase inhibitor homolog prevents cold-induced sweetening of potato tubers. Nat Biotechnol 17:708–711
Gupta R, Jung E, Brunak S (2004) Prediction of N-glycosylation sites in human proteins. http://www.cbs.dtu.dk/services/NetNGlyc
Holland JB, Helland SJ, Sharopova N, Rhyne DC (2001) Polymorphism of PCR-based markers targeting exons, introns, promoter regions, and SSRs in maize and introns and repeat sequences in oat. Genome 44:1065–1076
Hothorn M, D’Angelo I, Marquez JA, Greiner S, Scheffzek K (2004a) The invertase inhibitor Nt-CIF from tobacco: a highly thermostable four-helix bundle with an unusual N-terminal extension. J Mol Biol 335:987–995
Hothorn M, Wolf S, Aloy P, Greiner S, Scheffzek K (2004b) Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins. Plant Cell 16:3437–3447
Hothorn M, Van den Ende W, Lammens W, Rybin V, Scheffzek K (2010) Structural insights into the pH-controlled targeting of plant cell-wall invertase by a specific inhibitor protein. Proc Natl Acad Sci USA 107:17427–17432
Jehan T, Lakhanpaul S (2006) Single nucleotide polymorphism (SNP)—methods and applications in plant genetics: a review. Indian J Biotech 5:435–459
Krausgrill S, Greiner S, Koster U, Vogel R, Rausch T (1998) In transformed tobacco cells the apoplasmic invertase inhibitor operates as a regulatory switch of cell wall invertase. Plant J 13:275–280
Le Hir H, Nott A, Moore MJ (2003) How introns influence and enhance gene expression. TiBS 28:215–220
Liu X, Song B, Zhang H, Li X-Q, Xie C, Liu J (2010) Cloning and molecular characterization of putative invertase inhibitor genes and their possible contributions to cold-induced sweetening of potato tubers. Mol Genet Genomics 284:147–159
Pressey R (1966) Separation and properties of potato invertase and invertase inhibitor. Arch Biochem Biophys 113:667–674
Pressey R (1967) Invertase inhibitor from potatoes: purification, characterization, and reactivity with plant invertases. Plant Physiol 42:1780–1786
Pressey R (1968) Invertase inhibitors from red beet, sugar beet, and sweet potato roots. Plant Physiol 43:1430–1434
Pressey R (1994) Invertase inhibitor in tomato fruit. Phytochemistry 36:543–546
Rausch T, Greiner S (2004) Plant protein inhibitors of invertases. Biochim Biophys Acta 1696:253–261
Reca IB, Brutus A, D’Avino R, Villard C, Bellincampim D, Giardina T (2008) Molecular cloning, expression and characterization of a novel apoplastic invertase inhibitor from tomato (Solanum lycopersicum) and its use to purify a vacuolar invertase. Biochimie 90:1611–1623
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology, Humana Press, Totowa, pp 365–386
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Sander A, Krausgrill S, Greiner S, Weil M, Rausch T (1996) Sucrose protects cell wall invertase but not vacuolar invertase against proteinaceous inhibitors. FEBS Lett 385:171–175
Schwimmer S, Makower RU, Rorem ES (1961) Invertase & invertase inhibitor in potato. Plant Physiol 36:313–316
Simko I (2004) One potato, two potato: haplotype association mapping in autotetraploids. Trends Plant Sci 9:441–448
Simko I, Haynes KG, Jones RW (2006) Assessment of linkage disequilibrium in potato genome with single nucleotide polymorphism markers. Genetics 173:2237–2245
Staden R, Beal KF, Bonfield JK (1999) The Staden package, 1998. Methods Mol Biol 132:115–130
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tang G-Q, Lüscher M, Sturm A (1999) Antisense repression of vacuolar and cell wall invertase in transgenic carrot alters early plant development and sucrose partitioning. Plant Cell 11:177–189
The Potato Genome Sequencing Consortium (2011) Genome sequence and analysis of the tuber crop potato. Nature 475:189–195
von Heijne G (1984) How signal sequences maintain cleavage specificity. J Mol Biol 173:243–251
von Heijne G (1985) Signal sequences: the limits of variation. J Mol Biol 184:99–105
Weil M, Krausgrill S, Schuster A, Rausch T (1994) A 17-KDa Nicotiana tabacum cell-wall peptide acts as an in vitro inhibitor of the cell-wall isoform of acid invertase. Planta 193:438–445
Acknowledgments
We would like to thank Russell Genet for supplying potato tubers, Samantha Baldwin for help in primer design, and Mei Meiyalaghan and Mark Fiers for stimulating discussions. This work was supported by Ministry of Science & Innovation (New Zealand) contract C02X0805 to The New Zealand Institute for Plant & Food Research Limited, New Zealand, and grants from Lincoln University, New Zealand (SSD) and a New Zealand International Doctoral Research Scholarship (SSD).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Gebhardt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig 1 Alignment of the genomic sequences of apoplastic invertase inhibitor alleles. Comparative alignment of the genomic sequences of the apoplastic invertase inhibitor alleles identified in the present study with sequences of apoplastic invertase inhibitors previously isolated from potato. ‘DM1-3 516 R44′ is superscaffold PGSC0003DMB000000114 (position 619958 to 618056) from the potato genome sequence (The Potato Genome Sequencing Consortium, 2011); Stinvinh1 (GU321338) and Stinvinh3 (GU321339) (Liu et al. 2010). Primer sites are highlighted in yellow with forward and reverse arrows, exons are in bold with a red box around the start and stop codons, substitutions/indels are shaded gray, “-“indicates a deletion or absence of sequence information. The conserved 5′GT and 3′AG dinucleotides at the exon/intron boundaries are indicated by blue arrows. Predicted changes in the amino acid sequence are presented in their one letter code below the corresponding underlined nucleotide codon. SumDel = ‘Summer Delight’.
Supplementary Table 1Nucleotide polymorphism in the putative mRNA of invertase inhibitor alleles. Nucleotide sequence and amino acid differences in the predicted mRNA sequences of invertase inhibitor alleles compared with the allele inh1-a (GenBank accession number AY864819). Only polymorphisms in the exons are presented. SNPs in StInvInh3 at nt 78 (C to T) and inh1-c at nt 266 (A to G) do not cause a change in amino acid. The numbering is as in Supplementary Figure 1. StInvInh1 and StInvInh3, Liu et al 2010.
Rights and permissions
About this article
Cite this article
Datir, S.S., Latimer, J.M., Thomson, S.J. et al. Allele diversity for the apoplastic invertase inhibitor gene from potato. Mol Genet Genomics 287, 451–460 (2012). https://doi.org/10.1007/s00438-012-0690-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-012-0690-z